Uncovering the significance of expanded CD8+ large granular lymphocytes in inclusion body myositis: Insights into T cell phenotype and functional alterations, and disease severity
- PMID: 37063893
- PMCID: PMC10098158
- DOI: 10.3389/fimmu.2023.1153789
Uncovering the significance of expanded CD8+ large granular lymphocytes in inclusion body myositis: Insights into T cell phenotype and functional alterations, and disease severity
Abstract
Introduction: Inclusion body myositis (IBM) is a progressive inflammatory myopathy characterised by skeletal muscle infiltration and myofibre invasion by CD8+ T lymphocytes. In some cases, IBM has been reported to be associated with a systemic lymphoproliferative disorder of CD8+ T cells exhibiting a highly differentiated effector phenotype known as T cell Large Granular Lymphocytic Leukemia (T-LGLL).
Methods: We investigated the incidence of a CD8+ T-LGL lymphoproliferative disorder in 85 IBM patients and an aged-matched group of 56 Healthy Controls (HC). Further, we analysed the phenotypical characteristics of the expanded T-LGLs and investigated whether their occurrence was associated with any particular HLA alleles or clinical characteristics.
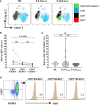
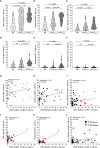
Results: Blood cell analysis by flow cytometry revealed expansion of T-LGLs in 34 of the 85 (40%) IBM patients. The T cell immunophenotype of T-LGLHIGH patients was characterised by increased expression of surface molecules including CD57 and KLRG1, and to a lesser extent of CD94 and CD56 predominantly in CD8+ T cells, although we also observed modest changes in CD4+ T cells and γδ T cells. Analysis of Ki67 in CD57+ KLRG1+ T cells revealed that only a small proportion of these cells was proliferating. Comparative analysis of CD8+ and CD4+ T cells isolated from matched blood and muscle samples donated by three patients indicated a consistent pattern of more pronounced alterations in muscles, although not significant due to small sample size. In the T-LGLHIGH patient group, we found increased frequencies of perforin-producing CD8+ and CD4+ T cells that were moderately correlated to combined CD57 and KLRG1 expression. Investigation of the HLA haplotypes of 75 IBM patients identified that carriage of the HLA-C*14:02:01 allele was significantly higher in T-LGLHIGH compared to T-LGLLOW individuals. Expansion of T-LGL was not significantly associated with seropositivity patient status for anti-cytosolic 5'-nucleotidase 1A autoantibodies. Clinically, the age at disease onset and disease duration were similar in the T-LGLHIGH and T-LGLLOW patient groups. However, metadata analysis of functional alterations indicated that patients with expanded T-LGL more frequently relied on mobility aids than T-LGLLOW patients indicating greater disease severity.
Conclusion: Altogether, these results suggest that T-LGL expansion occurring in IBM patients is correlated with exacerbated immune dysregulation and increased disease burden.
Keywords: KLRG1; T cell large granular lymphocytes; inclusion body myositis; inhibitory natural killer receptors; senescence.
Copyright © 2023 McLeish, Sooda, Slater, Kachigunda, Beer, Paramalingam, Lamont, Chopra, Mastaglia, Needham and Coudert.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Goyal NA, Cash TM, Alam U, Enam S, Tierney P, Araujo N, et al. . Seropositivity for Nt5c1a antibody in sporadic inclusion body myositis predicts more severe motor, bulbar and respiratory involvement. J Neurol Neurosurg Psychiatry (2016) 87(4):373–8. doi: 10.1136/jnnp-2014-310008 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

