HSF1 promotes CD69+ Treg differentiation to inhibit colitis progression
- PMID: 37064870
- PMCID: PMC10091886
- DOI: 10.7150/thno.78078
HSF1 promotes CD69+ Treg differentiation to inhibit colitis progression
Abstract
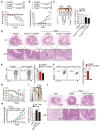
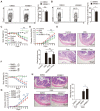
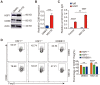
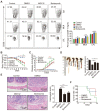
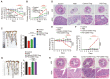
Regulatory T cells (Tregs) are critical for generating and maintaining peripheral tolerance. Treg-based immunotherapy is valuable for the clinical management of diseases resulting from dysregulation of immune tolerance. However, the lack of potency is a potential limitation of Treg therapy. In addition, CD69 positive-Treg (CD69+ Treg) represent a newly identified subset of Tregs with potent immune suppressive capability. Methods: Foxp3 YFP-Cre CD69 fl/fl and CD4 Cre CD69 fl/fl mice were generated to determine the relevance of CD69 to Treg. Chromatin Immunoprecipitation Assay (ChIP) and luciferase Assay were performed to detect the regulation of CD69 transcription by heat shock transcription factor 1(HSF1). Gene expression was measured by western blotting and qRT-PCR. The differentiation of naive T cells to CD69+Foxp3+ iTregs was determined by flow cytometry. The immunosuppressive ability of Tregs was analyzed by ELISA and flow cytometry. Colon inflammation in mice was reflected by changes in body weight and colon length, the disease activity index (DAI), and H&E staining of colon tissues. Results: Induced Tregs (iTregs) from CD4 Cre CD69 fl/fl mice failed to alleviate colitis. The transcription factor HSF1 interacted with the promoter of the CD69 gene to prompt its transcription during Treg differentiation. Genetic and chemical inhibition of HSF1 impaired CD69+ Treg differentiation and promoted the pathogenesis of colitis in mice. In contrast, HSF1 protein stabilized by inhibiting its proteasomal degradation promoted CD69+ Treg differentiation and alleviated colitis in mice. Moreover, adoptive transfer of iTregs with HSF1 stabilization by proteasome inhibitor (PSI) dramatically prevented the development of colitis in mice and was accompanied by decreased production of pro-inflammatory cytokines and reduced accumulation of pro-inflammatory lymphocytes in colitis tissue, whereas Tregs induced in the absence of PSI were less stable and ineffective in suppressing colitis. Conclusions: HSF1 promotes CD69+ Tregs differentiation by activating the CD69 transcription, which is critical for the immunosuppressive function of Tregs. Stabilization of HSF1 by PSIs results in the efficient generation of Tregs with high potency to treat colitis and probably other autoimmune diseases involving Tregs deficiency.
Keywords: CD69; HSF1; IBD; Treg; proteasome inhibitor.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


Similar articles
-
Heat shock factor 1 drives regulatory T-cell induction to limit murine intestinal inflammation.Mucosal Immunol. 2024 Feb;17(1):94-110. doi: 10.1016/j.mucimm.2023.11.003. Epub 2023 Nov 7. Mucosal Immunol. 2024. PMID: 37944754 Free PMC article.
-
Regulatory role of PI16 in autoimmune arthritis and intestinal inflammation: implications for Treg cell differentiation and function.J Transl Med. 2024 Apr 2;22(1):327. doi: 10.1186/s12967-024-05082-1. J Transl Med. 2024. PMID: 38566233 Free PMC article.
-
CD69 enhances immunosuppressive function of regulatory T-cells and attenuates colitis by prompting IL-10 production.Cell Death Dis. 2018 Sep 5;9(9):905. doi: 10.1038/s41419-018-0927-9. Cell Death Dis. 2018. PMID: 30185773 Free PMC article.
-
Foxp3, Regulatory T Cell, and Autoimmune Diseases.Inflammation. 2017 Feb;40(1):328-339. doi: 10.1007/s10753-016-0470-8. Inflammation. 2017. PMID: 27882473 Review.
-
Treg plasticity and human diseases.Inflamm Res. 2023 Dec;72(12):2181-2197. doi: 10.1007/s00011-023-01808-x. Epub 2023 Oct 25. Inflamm Res. 2023. PMID: 37878023 Review.
Cited by
-
CD69 is a Promising Immunotherapy and Prognosis Prediction Target in Cancer.Immunotargets Ther. 2024 Jan 9;13:1-14. doi: 10.2147/ITT.S439969. eCollection 2024. Immunotargets Ther. 2024. PMID: 38223406 Free PMC article. Review.
-
Artesunate alleviated murine ulcerative colitis by regulating immune response through inhibiting endoplasmic reticulum stress.Front Immunol. 2025 Feb 26;16:1545468. doi: 10.3389/fimmu.2025.1545468. eCollection 2025. Front Immunol. 2025. PMID: 40079012 Free PMC article.
-
LINC00942 inhibits ferroptosis and induces the immunosuppression of regulatory T cells by recruiting IGF2BP3/SLC7A11 in hepatocellular carcinoma.Funct Integr Genomics. 2024 Feb 14;24(1):29. doi: 10.1007/s10142-024-01292-4. Funct Integr Genomics. 2024. PMID: 38353724 Free PMC article.
-
HSF1 Activation Mechanisms, Disease Roles, and Small Molecule Therapeutics.Int J Biol Sci. 2025 Apr 28;21(8):3351-3378. doi: 10.7150/ijbs.110447. eCollection 2025. Int J Biol Sci. 2025. PMID: 40520012 Free PMC article. Review.
-
CD69 Expression is Negatively Associated With T-Cell Immunity and Predicts Antiviral Therapy Response in Chronic Hepatitis B.Ann Lab Med. 2025 Mar 1;45(2):185-198. doi: 10.3343/alm.2024.0178. Epub 2024 Dec 20. Ann Lab Med. 2025. PMID: 39703148 Free PMC article.
References
-
- Aharoni R, Sonego H, Brenner O, Eilam R, Arnon R. The therapeutic effect of glatiramer acetate in a murine model of inflammatory bowel disease is mediated by anti-inflammatory T-cells. Immunol Lett. 2007;112(2):110–119. - PubMed
-
- Dickson I. IBD: Ustekinumab therapy for Crohn's disease. Nat Rev Gastroenterol Hepatol. 2017;14(1):4. - PubMed
-
- Kitagawa Y, Sakaguchi S. Molecular control of regulatory T cell development and function. Curr Opin Immunol. 2017;49:64–70. - PubMed
-
- Tanoue T, Atarashi K, Honda K. Development and maintenance of intestinal regulatory T cells. Nat Rev Immunol. 2016;16(5):295–309. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

