Identification of Novel Tau-Tubulin Kinase 2 Inhibitors Using Computational Approaches
- PMID: 37065061
- PMCID: PMC10099139
- DOI: 10.1021/acsomega.3c00225
Identification of Novel Tau-Tubulin Kinase 2 Inhibitors Using Computational Approaches
Abstract
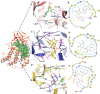
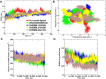
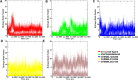
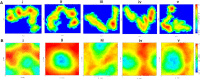
Tau tubulin kinase 2 (TTBK2) associated with multiple diseases is one of the kinases which phosphorylates tau and tubulin. Numerous efforts have been made to understand the role of TTBK2 in protein folding mechanisms and misfolding behavior. The misfolded protein intermediates form polymers with unwanted aggregation properties that initiate several diseases, including Alzheimer's. The availability of TTBK2 inhibitors can enhance the understanding of the molecular mechanism of action of the kinase and assist in developing novel therapeutics. In the quest for TTBK2 inhibitors, this study focuses on screening two chemical libraries (ChEMBL and ZINC-FDA). The molecular docking, RO5/absorption, distribution, metabolism, and excretion/toxicity, density functional theory, molecular dynamics (MD) simulations, and molecular mechanics with generalized Born and surface area solvation techniques enabled shortlisting of the four most active compounds, namely, ChEMBL1236395, ChEMBL2104398, ChEMBL3427435, and ZINC000000509440. Moreover, 500 ns MD simulation was performed for each complex, which provided valuable insights into the structural changes in the complexes. The relative fluctuation, solvent accessible surface area, atomic gyration, compactness covariance, and free energy landscapes revealed that the compounds could stabilize the TTBK2 protein. Overall, this study would be valuable for the researchers targeting the development of novel TTBK2 inhibitors.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
A molecular journey to check the conformational dynamics of tau tubulin kinase 2 mutations associated with Alzheimer's disease.RSC Adv. 2021 Jan 5;11(3):1320-1331. doi: 10.1039/d0ra07659g. eCollection 2021 Jan 4. RSC Adv. 2021. PMID: 35424125 Free PMC article.
-
Computational Design of Novel Tau-Tubulin Kinase 1 Inhibitors for Neurodegenerative Diseases.Pharmaceuticals (Basel). 2024 Jul 16;17(7):952. doi: 10.3390/ph17070952. Pharmaceuticals (Basel). 2024. PMID: 39065802 Free PMC article.
-
Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.Sci Rep. 2023 Apr 14;13(1):6118. doi: 10.1038/s41598-023-32854-4. Sci Rep. 2023. PMID: 37059819 Free PMC article.
-
TTBK2: a tau protein kinase beyond tau phosphorylation.Biomed Res Int. 2015;2015:575170. doi: 10.1155/2015/575170. Epub 2015 Apr 9. Biomed Res Int. 2015. PMID: 25950000 Free PMC article. Review.
-
Tau-tubulin kinase.Front Mol Neurosci. 2014 Apr 28;7:33. doi: 10.3389/fnmol.2014.00033. eCollection 2014. Front Mol Neurosci. 2014. PMID: 24808823 Free PMC article. Review.
Cited by
-
Design, synthesis, and pharmacological evaluation of indazole carboxamides of N-substituted pyrrole derivatives as soybean lipoxygenase inhibitors.Mol Divers. 2024 Dec;28(6):3757-3782. doi: 10.1007/s11030-023-10775-8. Epub 2023 Dec 25. Mol Divers. 2024. PMID: 38145424
-
Paeonol Attenuates Atherosclerosis by Inhibiting Vascular Smooth Muscle Cells Senescence via SIRT1/P53/TRF2 Signaling Pathway.Molecules. 2024 Jan 4;29(1):261. doi: 10.3390/molecules29010261. Molecules. 2024. PMID: 38202844 Free PMC article.
-
Mechanistic insights into MARK4 inhibition by galantamine toward therapeutic targeting of Alzheimer's disease.Front Pharmacol. 2023 Sep 19;14:1276179. doi: 10.3389/fphar.2023.1276179. eCollection 2023. Front Pharmacol. 2023. PMID: 37795023 Free PMC article.
-
The identification of c-Abl inhibitors as potential agents for Parkinson's disease: a preliminary in silico approach.Mol Divers. 2024 Dec;28(6):4051-4065. doi: 10.1007/s11030-023-10796-3. Epub 2024 Jan 25. Mol Divers. 2024. PMID: 38273156
-
Synthesis, Characterization, Antiglycation Evaluation, Molecular Docking, and ADMET Studies of 4-Thiazolidinone Derivatives.ACS Omega. 2023 Dec 28;9(1):1810-1820. doi: 10.1021/acsomega.3c08463. eCollection 2024 Jan 9. ACS Omega. 2023. PMID: 38222574 Free PMC article.
References
-
- Watanabe T.; Kakeno M.; Matsui T.; Sugiyama I.; Arimura N.; Matsuzawa K.; Shirahige A.; Ishidate F.; Nishioka T.; Taya S.; Hoshino M.; Kaibuchi K. TTBK2 with EB1/3 regulates microtubule dynamics in migrating cells through KIF2A phosphorylation. J. Cell Biol. 2015, 210, 737–751. 10.1083/jcb.201412075. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

