Genome-resolved analyses of oligotrophic groundwater microbial communities along phenol pollution in a continuous-flow biodegradation model system
- PMID: 37065124
- PMCID: PMC10090433
- DOI: 10.3389/fmicb.2023.1147162
Genome-resolved analyses of oligotrophic groundwater microbial communities along phenol pollution in a continuous-flow biodegradation model system
Abstract
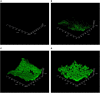
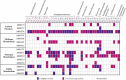
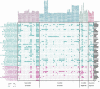
Groundwater pollution is one of the major environmental concerns. The entrance of pollutants into the oligotrophic groundwater ecosystems alters native microbial community structure and metabolism. This study investigated the application of innovative Small Bioreactor Chambers and CaO2 nanoparticles for phenol removal within continuous-flow sand-packed columns for 6 months. Scanning electron microscopy and confocal laser scanning microscopy analysis were conducted to indicate the impact of attached biofilm on sand surfaces in bioremediation columns. Then, the influence of each method on the microbial biodiversity of the column's groundwater was investigated by next-generation sequencing of the 16S rRNA gene. The results indicated that the simultaneous application of biostimulation and bioaugmentation completely eliminated phenol during the first 42 days. However, 80.2% of phenol remained in the natural bioremediation column at the end of the experiment. Microbial diversity was decreased by CaO2 injection while order-level groups known for phenol degradation such as Rhodobacterales and Xanthomonadales dominated in biostimulation columns. Genome-resolved comparative analyses of oligotrophic groundwater prokaryotic communities revealed that Burkholderiales, Micrococcales, and Cytophagales were the dominant members of the pristine groundwater. Six-month exposure of groundwater to phenol shifted the microbial population towards increasing the heterotrophic members of Desulfobacterales, Pseudomonadales, and Xanthomonadales with the degradation potential of phenol and other hydrocarbons.
Keywords: biodiversity; bioremediation; metagenome; oligotrophic groundwater; phenol.
Copyright © 2023 Yavari-Bafghi, Rezaei Somee, Amoozegar, Dastgheib and Shavandi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Abbasian F., Lockington R., Mallavarapu M., Naidu R. (2015). A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria. Appl. Biochem. Biotechnol. 176 670–699. - PubMed
-
- Abbasian F., Lockington R., Megharaj M., Naidu R. (2016). A review on the genetics of aliphatic and aromatic hydrocarbon degradation. Appl. Biochem. Biotechnol. 178 224–250. - PubMed
-
- Brescia F., Pertot I., Puopolo G. (2020). “Lysobacter,” in Beneficial Microbes in Agro-Ecology, eds Amaresan N., Senthil Kumar M., Annapurna K. (Amsterdam: Elsevier; ).
-
- Bushnell B. (2014). BBMap: a Fast, Accurate, Splice-Aware Aligner. Berkeley, CA: Lawrence Berkeley National Laboratory.
LinkOut - more resources
Full Text Sources

