Proteomic analysis of metronidazole resistance in the human facultative pathogen Bacteroides fragilis
- PMID: 37065137
- PMCID: PMC10102347
- DOI: 10.3389/fmicb.2023.1158086
Proteomic analysis of metronidazole resistance in the human facultative pathogen Bacteroides fragilis
Abstract
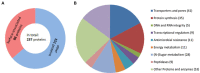
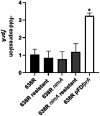
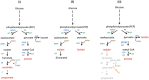
The anaerobic gut bacteria and opportunistic pathogen Bacteroides fragilis can cause life-threatening infections when leaving its niche and reaching body sites outside of the gut. The antimicrobial metronidazole is a mainstay in the treatment of anaerobic infections and also highly effective against Bacteroides spp. Although resistance rates have remained low in general, metronidazole resistance does occur in B. fragilis and can favor fatal disease outcomes. Most metronidazole-resistant Bacteroides isolates harbor nim genes, commonly believed to encode for nitroreductases which deactivate metronidazole. Recent research, however, suggests that the mode of resistance mediated by Nim proteins might be more complex than anticipated because they affect the cellular metabolism, e.g., by increasing the activity of pyruvate:ferredoxin oxidoreductase (PFOR). Moreover, although nim genes confer only low-level metronidazole resistance to Bacteroides, high-level resistance can be much easier induced in the laboratory in the presence of a nim gene than without. Due to these observations, we hypothesized that nim genes might induce changes in the B. fragilis proteome and performed comparative mass-spectrometric analyses with B. fragilis 638R, either with or without the nimA gene. Further, we compared protein expression profiles in both strains after induction of high-level metronidazole resistance. Interestingly, only few proteins were repeatedly found to be differentially expressed in strain 638R with the nimA gene, one of them being the flavodiiron protein FprA, an enzyme involved in oxygen scavenging. After induction of metronidazole resistance, a far higher number of proteins were found to be differentially expressed in 638R without nimA than in 638R with nimA. In the former, factors for the import of hemin were strongly downregulated, indicating impaired iron import, whereas in the latter, the observed changes were not only less numerous but also less specific. Both resistant strains, however, displayed a reduced capability of scavenging oxygen. Susceptibility to metronidazole could be widely restored in resistant 638R without nimA by supplementing growth media with ferrous iron sulfate, but not so in resistant 638R with the nimA gene. Finally, based on the results of this study, we present a novel hypothetic model of metronidazole resistance and NimA function.
Keywords: Bacteroides fragilis; antimicrobial resistance; metronidazole; nimA gene; proteomics.
Copyright © 2023 Paunkov, Hummel, Strasser, Sóki and Leitsch.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Modulation of Iron Import and Metronidazole Resistance in Bacteroides fragilis Harboring a nimA Gene.Front Microbiol. 2022 Jun 9;13:898453. doi: 10.3389/fmicb.2022.898453. eCollection 2022. Front Microbiol. 2022. PMID: 35756037 Free PMC article.
-
Initial expression levels of nimA are decisive for protection against metronidazole in Bacteroides fragilis.Anaerobe. 2022 Oct;77:102630. doi: 10.1016/j.anaerobe.2022.102630. Epub 2022 Aug 24. Anaerobe. 2022. PMID: 36028117
-
Inducible metronidazole resistance in nim-positive and nim-negative bacteroides fragilis group strains after several passages metronidazole containing columbia agar plates.Infection. 2005 Oct;33(5-6):368-72. doi: 10.1007/s15010-005-5061-9. Infection. 2005. PMID: 16258869
-
Metronidazole resistance and nim genes in anaerobes: A review.Anaerobe. 2019 Feb;55:40-53. doi: 10.1016/j.anaerobe.2018.10.004. Epub 2018 Oct 11. Anaerobe. 2019. PMID: 30316817 Review.
-
Mechanisms of Bacteroides fragilis resistance to metronidazole.Infect Genet Evol. 2018 Oct;64:156-163. doi: 10.1016/j.meegid.2018.06.020. Epub 2018 Jun 21. Infect Genet Evol. 2018. PMID: 29936037 Review.
Cited by
-
Multi-omics approach for understanding the response of Bacteroides fragilis to carbapenems.Heliyon. 2024 Aug 30;10(17):e37049. doi: 10.1016/j.heliyon.2024.e37049. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39286136 Free PMC article.
-
New functions of pirin proteins and a 2-ketoglutarate: Ferredoxin oxidoreductase ortholog in Bacteroides fragilis metabolism and their impact on antimicrobial susceptibility to metronidazole and amixicile.Microbiologyopen. 2024 Aug;13(4):e1429. doi: 10.1002/mbo3.1429. Microbiologyopen. 2024. PMID: 39109824 Free PMC article.
-
Hemophore-like proteins of the HmuY family in the oral and gut microbiome: unraveling the mystery of their evolution.Microbiol Mol Biol Rev. 2024 Mar 27;88(1):e0013123. doi: 10.1128/mmbr.00131-23. Epub 2024 Feb 2. Microbiol Mol Biol Rev. 2024. PMID: 38305743 Free PMC article. Review.
-
Proteomics-Based RT-qPCR and Functional Analysis of 18 Genes in Metronidazole Resistance of Bacteroides fragilis.Antibiotics (Basel). 2024 Feb 22;13(3):207. doi: 10.3390/antibiotics13030207. Antibiotics (Basel). 2024. PMID: 38534642 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

