Clostridioides difficile minimal nutrient requirements for flagellar motility
- PMID: 37065145
- PMCID: PMC10098170
- DOI: 10.3389/fmicb.2023.1172707
Clostridioides difficile minimal nutrient requirements for flagellar motility
Abstract
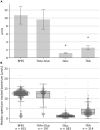
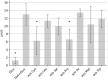
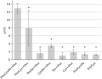
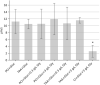
As many gastro-intestinal pathogens, the majority of Clostridioides difficile strains express flagella together with a complete chemotaxis system. The resulting swimming motility is likely contributing to the colonization success of this important pathogen. In contrast to the well investigated general energy metabolism of C. difficile, little is known about the metabolic requirements for maintaining the ion motive force across the membrane, which in turn powers the flagellar motor. We studied here systematically the effect of various amino acids and carbohydrates on the swimming velocity of C. difficile using video microscopy in conjunction with a software based quantification of the swimming speed. Removal of individual amino acids from the medium identified proline and cysteine as the most important amino acids that power swimming motility. Glycine, which is as proline one of the few amino acids that are reduced in Stickland reactions, was not critical for swimming motility. This suggests that the ion motive force that powers the flagellar motor, is critically depending on proline reduction. A maximal and stable swimming motility was achieved with only four compounds, including the amino acids proline, cysteine and isoleucine together with a single, but interchangeable carbohydrate source such as glucose, succinate, mannose, ribose, pyruvate, trehalose, or ethanolamine. We expect that the identified "minimal motility medium" will be useful in future investigations on the flagellar motility and chemotactic behavior in C. difficile, particularly for the unambiguous identification of chemoattractants.
Keywords: Clostridioides difficile; flagella; minimal medium optimization; motility; nutrients.
Copyright © 2023 Schwanbeck, Oehmig, Groß and Bohne.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
d-Proline Reductase Underlies Proline-Dependent Growth of Clostridioides difficile.J Bacteriol. 2022 Aug 16;204(8):e0022922. doi: 10.1128/jb.00229-22. Epub 2022 Jul 13. J Bacteriol. 2022. PMID: 35862761 Free PMC article.
-
The Stickland Reaction Precursor trans-4-Hydroxy-l-Proline Differentially Impacts the Metabolism of Clostridioides difficile and Commensal Clostridia.mSphere. 2022 Apr 27;7(2):e0092621. doi: 10.1128/msphere.00926-21. Epub 2022 Mar 30. mSphere. 2022. PMID: 35350846 Free PMC article.
-
Clostridioides difficile Single Cell Swimming Strategy: A Novel Motility Pattern Regulated by Viscoelastic Properties of the Environment.Front Microbiol. 2021 Jul 21;12:715220. doi: 10.3389/fmicb.2021.715220. eCollection 2021. Front Microbiol. 2021. PMID: 34367119 Free PMC article.
-
Food for thought-The link between Clostridioides difficile metabolism and pathogenesis.PLoS Pathog. 2023 Jan 5;19(1):e1011034. doi: 10.1371/journal.ppat.1011034. eCollection 2023 Jan. PLoS Pathog. 2023. PMID: 36602960 Free PMC article. Review.
-
Metabolism the Difficile Way: The Key to the Success of the Pathogen Clostridioides difficile.Front Microbiol. 2019 Feb 15;10:219. doi: 10.3389/fmicb.2019.00219. eCollection 2019. Front Microbiol. 2019. PMID: 30828322 Free PMC article. Review.
Cited by
-
Clostridioides difficile Flagella.Int J Mol Sci. 2024 Feb 12;25(4):2202. doi: 10.3390/ijms25042202. Int J Mol Sci. 2024. PMID: 38396876 Free PMC article. Review.
References
-
- Armitage J. P., Berry R. M. (2020). Assembly and dynamics of the bacterial flagellum. Annu. Rev. Microbiol. 74 181–200. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

