Causal effects of gut microbiota on the risk of chronic kidney disease: a Mendelian randomization study
- PMID: 37065213
- PMCID: PMC10102584
- DOI: 10.3389/fcimb.2023.1142140
Causal effects of gut microbiota on the risk of chronic kidney disease: a Mendelian randomization study
Abstract
Background: Previous studies have reported that gut microbiota is associated with an increased risk of chronic kidney disease (CKD) progression. However, whether gut microbiota has a causal effect on the development of CKD has not been revealed. Thus, we aimed to analyze the potential causal effect of gut microbiota on the risk of CKD using mendelian randomization (MR) study.
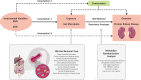
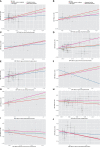
Materials and methods: Independent single nucleotide polymorphisms closely associated with 196 gut bacterial taxa (N = 18340) were identified as instrumental variables. Two-sample MR was performed to evaluate the causal effect of gut microbiota on CKD (N = 480698), including inverse-variance-weighted (IVW) method, weighted median method, MR-Egger, mode-based estimation and MR-PRESSO. The robustness of the estimation was tested by a series of sensitivity analyses including Cochran's Q test, MR-Egger intercept analysis, leave-one-out analysis and funnel plot. Statistical powers were also calculated.
Results: The genetically predicted higher abundance of order Desulfovibrionales was causally associated with an increased risk of CKD (odds ratio = 1.15, 95% confidence interval: 1.05-1.26; p = 0.0026). Besides, we also detected potential causalities between nine other taxa (Eubacterium eligens group, Desulfovibrionaceae, Ruminococcaceae UCG-002, Deltaproteobacteria, Lachnospiraceae UCG-010, Senegalimassilia, Peptostreptococcaceae, Alcaligenaceae and Ruminococcus torques group) and CKD (p < 0.05). No heterogeneity or pleiotropy was detected for significant estimates.
Conclusion: We found that Desulfovibrionales and nine other taxa are associated with CKD, thus confirming that gut microbiota plays an important role in the pathogenesis of CKD. Our work also provides new potential indicators and targets for screening and prevention of CKD.
Keywords: CKD; Mendelian randomization; chronic kidney disease; gut microbiota; nutrition.
Copyright © 2023 Luo, Cai, Luo, Hong, Xu, Lin, Chen and Fu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Genetic evidence supporting the causal role of gut microbiota in chronic kidney disease and chronic systemic inflammation in CKD: a bilateral two-sample Mendelian randomization study.Front Immunol. 2023 Nov 2;14:1287698. doi: 10.3389/fimmu.2023.1287698. eCollection 2023. Front Immunol. 2023. PMID: 38022507 Free PMC article.
-
Causal Effects of Specific Gut Microbiota on Chronic Kidney Diseases and Renal Function-A Two-Sample Mendelian Randomization Study.Nutrients. 2023 Jan 11;15(2):360. doi: 10.3390/nu15020360. Nutrients. 2023. PMID: 36678231 Free PMC article.
-
Causal effects of gut microbiota on appendicitis: a two-sample Mendelian randomization study.Front Cell Infect Microbiol. 2023 Dec 15;13:1320992. doi: 10.3389/fcimb.2023.1320992. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38162578 Free PMC article.
-
Genetic liability of gut microbiota for idiopathic pulmonary fibrosis and lung function: a two-sample Mendelian randomization study.Front Cell Infect Microbiol. 2024 May 22;14:1348685. doi: 10.3389/fcimb.2024.1348685. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38841114 Free PMC article.
-
Exploring the potential causal relationship between gut microbiota and heart failure: A two-sample mendelian randomization study combined with the geo database.Curr Probl Cardiol. 2024 Feb;49(2):102235. doi: 10.1016/j.cpcardiol.2023.102235. Epub 2023 Nov 30. Curr Probl Cardiol. 2024. PMID: 38040216 Review.
Cited by
-
Causal effects of gut microbiota on risk of overactive bladder symptoms: a two-sample Mendelian randomization study.Front Microbiol. 2024 Aug 23;15:1459634. doi: 10.3389/fmicb.2024.1459634. eCollection 2024. Front Microbiol. 2024. PMID: 39247701 Free PMC article.
-
Characterization of gut microbiota and metabolites in renal transplant recipients during COVID-19 and prediction of one-year allograft function.J Transl Med. 2025 Apr 10;23(1):420. doi: 10.1186/s12967-025-06090-5. J Transl Med. 2025. PMID: 40211390 Free PMC article.
-
Causal relationship between gut microbiota and male erectile dysfunction: a Mendelian randomization analysis.Front Microbiol. 2024 Aug 29;15:1367740. doi: 10.3389/fmicb.2024.1367740. eCollection 2024. Front Microbiol. 2024. PMID: 39268537 Free PMC article.
-
Mendelian Randomization Studies: Opening a New Window in the Study of Metabolic Diseases and Chronic Kidney Disease.Endocr Metab Immune Disord Drug Targets. 2025;25(6):442-457. doi: 10.2174/0118715303288685240808073238. Endocr Metab Immune Disord Drug Targets. 2025. PMID: 39171476 Review.
-
Causal role of immune cells in diabetic nephropathy: a bidirectional Mendelian randomization study.Front Endocrinol (Lausanne). 2024 Sep 13;15:1357642. doi: 10.3389/fendo.2024.1357642. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39345891 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

