A Complex of Pyrosequencing-Based Methods for Detection of Somatic Mutations in Codons 600 and 601 of the BRAF gene
- PMID: 37065428
- PMCID: PMC10090924
- DOI: 10.17691/stm2022.14.2.04
A Complex of Pyrosequencing-Based Methods for Detection of Somatic Mutations in Codons 600 and 601 of the BRAF gene
Abstract
The aim of the study is to develop methods for the differentiation of mutations in the BRAF codon 600 and to increase the sensitivity of the K601E mutation detection.
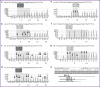
Materials and methods: The nucleotide sequence of the BRAF codons 592-602 was identified using the PyroMark Q24 genetic analysis system. The mutations search in codon 600 was conducted using the 600-S primer in line with the following order of adding nucleotides: GCTGTCАTCTGCTAGCTAGAC (corresponding to nucleotides 1799-1786). The K601E mutation was detected using the 601-S primer in line with the following order of nucleotide addition: GCTACTCACTGTAG (corresponding to nucleotides 1801-1793). The analytical characteristics of the proposed methods for somatic mutations' detection were determined using dilutions of plasmid DNA samples containing the BRAF gene region without mutations or with one of the following mutations: V600E, V600R, V600K, V600M, and K601E. Validation was performed on 132 samples of biological material obtained from the thyroid nodules.
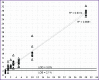
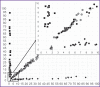
Results: The developed methods allow to determine 2% of the V600E or V600M mutations, 1% of the V600K and V600R mutations, and 3% of the K601E mutations in samples with high DNA concentration; it is also possible to confidently detect at least 5% of the mutant allele for all mutations in low concentration samples (less than 500 copies/PCR). During biological material testing, 53 samples with the V600E mutation were detected; the proportion of the mutant allele was 4.9-50.0%.
Conclusion: A complex of methods for determination of the nucleotide sequence of the BRAF codons 592-601 and the algorithm for testing samples and analyzing mutations in the BRAF codons 600-601 was developed. The method provides sufficient sensitivity to detect frequent mutations in codons 600 and 601 and allows them to be precisely differentiated.
Keywords: BRAF; fine-needle aspiration biopsy; oncogenetics; pyrosequencing; thyroid cancer.
Conflict of interest statement
Conflicts of interest. The authors declare that they have no conflicts of interest.
Figures
References
-
- Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E., Fish P., Harsha B., Hathaway C., Jupe S.C., Kok C.Y., Noble K., Ponting L., Ramshaw C.C., Rye C.E., Speedy H.E., Stefancsik R., Thompson S.L., Wang S., Ward S., Campbell P.J., Forbes S.A. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
