Clustering and machine learning-based integration identify cancer associated fibroblasts genes' signature in head and neck squamous cell carcinoma
- PMID: 37065499
- PMCID: PMC10098459
- DOI: 10.3389/fgene.2023.1111816
Clustering and machine learning-based integration identify cancer associated fibroblasts genes' signature in head and neck squamous cell carcinoma
Abstract
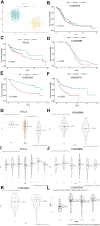
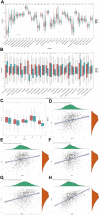
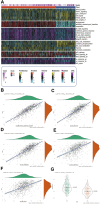
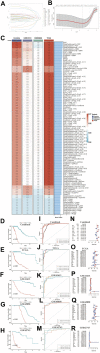
Background: A hallmark signature of the tumor microenvironment in head and neck squamous cell carcinoma (HNSCC) is abundantly infiltration of cancer-associated fibroblasts (CAFs), which facilitate HNSCC progression. However, some clinical trials showed targeted CAFs ended in failure, even accelerated cancer progression. Therefore, comprehensive exploration of CAFs should solve the shortcoming and facilitate the CAFs targeted therapies for HNSCC. Methods: In this study, we identified two CAFs gene expression patterns and performed the single-sample gene set enrichment analysis (ssGSEA) to quantify the expression and construct score system. We used multi-methods to reveal the potential mechanisms of CAFs carcinogenesis progression. Finally, we integrated 10 machine learning algorithms and 107 algorithm combinations to construct most accurate and stable risk model. The machine learning algorithms contained random survival forest (RSF), elastic network (Enet), Lasso, Ridge, stepwise Cox, CoxBoost, partial least squares regression for Cox (plsRcox), supervised principal components (SuperPC), generalised boosted regression modelling (GBM), and survival support vector machine (survival-SVM). Results: There are two clusters present with distinct CAFs genes pattern. Compared to the low CafS group, the high CafS group was associated with significant immunosuppression, poor prognosis, and increased prospect of HPV negative. Patients with high CafS also underwent the abundant enrichment of carcinogenic signaling pathways such as angiogenesis, epithelial mesenchymal transition, and coagulation. The MDK and NAMPT ligand-receptor cellular crosstalk between the cancer associated fibroblasts and other cell clusters may mechanistically cause immune escape. Moreover, the random survival forest prognostic model that was developed from 107 machine learning algorithm combinations could most accurately classify HNSCC patients. Conclusion: We revealed that CAFs would cause the activation of some carcinogenesis pathways such as angiogenesis, epithelial mesenchymal transition, and coagulation and revealed unique possibilities to target glycolysis pathways to enhance CAFs targeted therapy. We developed an unprecedentedly stable and powerful risk score for assessing the prognosis. Our study contributes to the understanding of the CAFs microenvironment complexity in patients with head and neck squamous cell carcinoma and serves as a basis for future in-depth CAFs gene clinical exploration.
Keywords: HNSCC; cancer-associated fibroblasts (CAFs); machine learning; prognosis; the tumor microenvironment.
Copyright © 2023 Wang, Zhao, Wang and Tan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Machine-learning derived identification of prognostic signature to forecast head and neck squamous cell carcinoma prognosis and drug response.Front Immunol. 2024 Dec 19;15:1469895. doi: 10.3389/fimmu.2024.1469895. eCollection 2024. Front Immunol. 2024. PMID: 39749326 Free PMC article.
-
Machine learning-based identification of a consensus immune-derived gene signature to improve head and neck squamous cell carcinoma therapy and outcome.Front Pharmacol. 2024 Apr 10;15:1341346. doi: 10.3389/fphar.2024.1341346. eCollection 2024. Front Pharmacol. 2024. PMID: 38666027 Free PMC article.
-
Identification and verification of eight cancer-associated fibroblasts related genes as a prognostic signature for head and neck squamous cell carcinoma.Heliyon. 2023 Feb 28;9(3):e14003. doi: 10.1016/j.heliyon.2023.e14003. eCollection 2023 Mar. Heliyon. 2023. PMID: 36938461 Free PMC article.
-
Cancer-associated fibroblasts: Mediators of head and neck tumor microenvironment remodeling.Biochim Biophys Acta Rev Cancer. 2023 Sep;1878(5):188940. doi: 10.1016/j.bbcan.2023.188940. Epub 2023 Jun 16. Biochim Biophys Acta Rev Cancer. 2023. PMID: 37331641 Review.
-
Cancer-Associated Fibroblast Density, Prognostic Characteristics, and Recurrence in Head and Neck Squamous Cell Carcinoma: A Meta-Analysis.Front Oncol. 2020 Nov 27;10:565306. doi: 10.3389/fonc.2020.565306. eCollection 2020. Front Oncol. 2020. PMID: 33330034 Free PMC article.
Cited by
-
Integrated Bioinformatics and Experimental Validation to Identify a Disulfidptosis-Related lncRNA Model for Prognostic Prediction in Papillary Renal Cell Carcinoma.Comb Chem High Throughput Screen. 2025;28(5):883-898. doi: 10.2174/0113862073303084240403051346. Comb Chem High Throughput Screen. 2025. PMID: 38639274
-
Creating a multifaceted prognostic model for cutaneous melanoma: the convergence of single-cell and bulk sequencing with machine learning.Front Cell Dev Biol. 2024 May 6;12:1401945. doi: 10.3389/fcell.2024.1401945. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38770150 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

