Spermiogenesis alterations in the absence of CTCF revealed by single cell RNA sequencing
- PMID: 37065848
- PMCID: PMC10097911
- DOI: 10.3389/fcell.2023.1119514
Spermiogenesis alterations in the absence of CTCF revealed by single cell RNA sequencing
Abstract
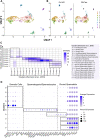
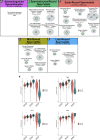
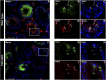
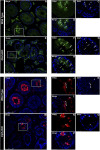
CTCF is an architectonic protein that organizes the genome inside the nucleus in almost all eukaryotic cells. There is evidence that CTCF plays a critical role during spermatogenesis as its depletion produces abnormal sperm and infertility. However, defects produced by its depletion throughout spermatogenesis have not been fully characterized. In this work, we performed single cell RNA sequencing in spermatogenic cells with and without CTCF. We uncovered defects in transcriptional programs that explain the severity of the damage in the produced sperm. In the early stages of spermatogenesis, transcriptional alterations are mild. As germ cells go through the specialization stage or spermiogenesis, transcriptional profiles become more altered. We found morphology defects in spermatids that support the alterations in their transcriptional profiles. Altogether, our study sheds light on the contribution of CTCF to the phenotype of male gametes and provides a fundamental description of its role at different stages of spermiogenesis.
Keywords: CTCF; ScRNA-seq; mouse testis; sperm; spermatogenesis.
Copyright © 2023 Torres-Flores, Díaz-Espinosa, López-Santaella, Rebollar-Vega, Vázquez-Jiménez, Taylor, Ortiz-Hernández, Echeverría, Vázquez-Nin, Gutierrez-Ruiz, De la Rosa-Velázquez, Resendis-Antonio and Hernández-Hernandez.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

