Assessing epidemic curves for evidence of superspreading
- PMID: 37066104
- PMCID: PMC10092342
- DOI: 10.1111/rssa.12919
Assessing epidemic curves for evidence of superspreading
Abstract
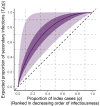
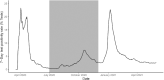
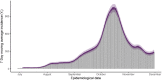
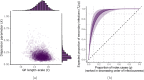
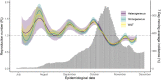
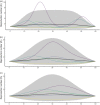
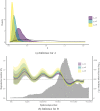
The expected number of secondary infections arising from each index case, referred to as the reproduction or number, is a vital summary statistic for understanding and managing epidemic diseases. There are many methods for estimating ; however, few explicitly model heterogeneous disease reproduction, which gives rise to superspreading within the population. We propose a parsimonious discrete-time branching process model for epidemic curves that incorporates heterogeneous individual reproduction numbers. Our Bayesian approach to inference illustrates that this heterogeneity results in less certainty on estimates of the time-varying cohort reproduction number . We apply these methods to a COVID-19 epidemic curve for the Republic of Ireland and find support for heterogeneous disease reproduction. Our analysis allows us to estimate the expected proportion of secondary infections attributable to the most infectious proportion of the population. For example, we estimate that the 20% most infectious index cases account for approximately 75%-98% of the expected secondary infections with 95% posterior probability. In addition, we highlight that heterogeneity is a vital consideration when estimating .
Keywords: branching processes; heterogeneous disease reproduction; time‐varying reproduction numbers.
© 2022 The Authors. Journal of the Royal Statistical Society: Series A (Statistics in Society) published by John Wiley & Sons Ltd on behalf of Royal Statistical Society.
Figures
Similar articles
-
A statistical framework for tracking the time-varying superspreading potential of COVID-19 epidemic.Epidemics. 2023 Mar;42:100670. doi: 10.1016/j.epidem.2023.100670. Epub 2023 Jan 24. Epidemics. 2023. PMID: 36709540 Free PMC article.
-
Chopping the tail: How preventing superspreading can help to maintain COVID-19 control.Epidemics. 2021 Mar;34:100430. doi: 10.1016/j.epidem.2020.100430. Epub 2020 Dec 21. Epidemics. 2021. PMID: 33360871 Free PMC article.
-
Chopping the tail: how preventing superspreading can help to maintain COVID-19 control.medRxiv [Preprint]. 2020 Jul 3:2020.06.30.20143115. doi: 10.1101/2020.06.30.20143115. medRxiv. 2020. Update in: Epidemics. 2021 Mar;34:100430. doi: 10.1016/j.epidem.2020.100430. PMID: 32637966 Free PMC article. Updated. Preprint.
-
International travel-related control measures to contain the COVID-19 pandemic: a rapid review.Cochrane Database Syst Rev. 2021 Mar 25;3(3):CD013717. doi: 10.1002/14651858.CD013717.pub2. Cochrane Database Syst Rev. 2021. PMID: 33763851 Free PMC article.
-
Travel-related control measures to contain the COVID-19 pandemic: a rapid review.Cochrane Database Syst Rev. 2020 Oct 5;10:CD013717. doi: 10.1002/14651858.CD013717. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Mar 25;3:CD013717. doi: 10.1002/14651858.CD013717.pub2. PMID: 33502002 Updated.
Cited by
-
Hybrid prediction of infections and deaths due to COVID-19 in two Colombian data series.PLoS One. 2023 Jun 8;18(6):e0286643. doi: 10.1371/journal.pone.0286643. eCollection 2023. PLoS One. 2023. PMID: 37289738 Free PMC article.
-
Accuracy of Inferences About the Reproductive Number and Superspreading Potential of SARS-CoV-2 with Incomplete Contact Tracing Data.Res Sq [Preprint]. 2023 Dec 29:rs.3.rs-3760127. doi: 10.21203/rs.3.rs-3760127/v1. Res Sq. 2023. PMID: 38234843 Free PMC article. Preprint.
References
-
- Anderson, R.M. & May, R.M. (1992) Infectious diseases of humans: dynamics and control. Oxford: Oxford University Press.
-
- Becker, N.G. & Britton, T. (1999) Statistical studies of infectious disease incidence. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 61(2), 287–307.
LinkOut - more resources
Full Text Sources
