This is a preprint.
Histone Deacetylases (HDACs) maintain expression of the pluripotent gene network via recruitment of RNA polymerase II to coding and non-coding loci
- PMID: 37066171
- PMCID: PMC10104071
- DOI: 10.1101/2023.04.06.535398
Histone Deacetylases (HDACs) maintain expression of the pluripotent gene network via recruitment of RNA polymerase II to coding and non-coding loci
Update in
-
Histone deacetylases maintain expression of the pluripotent gene network via recruitment of RNA polymerase II to coding and noncoding loci.Genome Res. 2024 Feb 7;34(1):34-46. doi: 10.1101/gr.278050.123. Genome Res. 2024. PMID: 38290976 Free PMC article.
Abstract
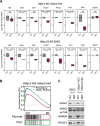
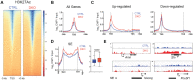
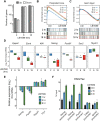
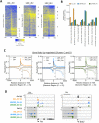
Histone acetylation is a dynamic modification regulated by the opposing actions of histone acetyltransferases (HATs) and histone deacetylases (HDACs). Deacetylation of histone tails results in chromatin tightening and therefore HDACs are generally regarded as transcriptional repressors. Counterintuitively, simultaneous deletion of Hdac1 and Hdac2 in embryonic stem cells (ESC) reduced expression of pluripotent transcription factors, Oct4, Sox2 and Nanog (OSN). By shaping global histone acetylation patterns, HDACs indirectly regulate the activity of acetyl-lysine readers, such as the transcriptional activator, BRD4. We used inhibitors of HDACs and BRD4 (LBH589 and JQ1 respectively) in combination with precision nuclear run-on and sequencing (PRO-seq) to examine their roles in defining the ESC transcriptome. Both LBH589 and JQ1 caused a marked reduction in the pluripotent network. However, while JQ1 treatment induced widespread transcriptional pausing, HDAC inhibition caused a reduction in both paused and elongating polymerase, suggesting an overall reduction in polymerase recruitment. Using enhancer RNA (eRNA) expression to measure enhancer activity we found that LBH589-sensitive eRNAs were preferentially associated with super-enhancers and OSN binding sites. These findings suggest that HDAC activity is required to maintain pluripotency by regulating the OSN enhancer network via the recruitment of RNA polymerase II.
Figures



References
-
- Arnold M, Bressin A, Jasnovidova O, Meierhofer D, Mayer A. 2021. A BRD4-mediated elongation control point primes transcribing RNA polymerase II for 3’-processing and termination. Mol Cell 81: 3589–3603 e3513. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
