This is a preprint.
SARS-CoV-2 Mac1 is required for IFN antagonism and efficient virus replication in mice
- PMID: 37066301
- PMCID: PMC10104158
- DOI: 10.1101/2023.04.06.535927
SARS-CoV-2 Mac1 is required for IFN antagonism and efficient virus replication in mice
Update in
-
SARS-CoV-2 Mac1 is required for IFN antagonism and efficient virus replication in cell culture and in mice.Proc Natl Acad Sci U S A. 2023 Aug 29;120(35):e2302083120. doi: 10.1073/pnas.2302083120. Epub 2023 Aug 22. Proc Natl Acad Sci U S A. 2023. PMID: 37607224 Free PMC article.
Abstract
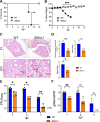
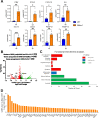
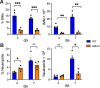
Several coronavirus (CoV) encoded proteins are being evaluated as targets for antiviral therapies for COVID-19. Included in this set of proteins is the conserved macrodomain, or Mac1, an ADP-ribosylhydrolase and ADP-ribose binding protein. Utilizing point mutant recombinant viruses, Mac1 was shown to be critical for both murine hepatitis virus (MHV) and severe acute respiratory syndrome (SARS)-CoV virulence. However, as a potential drug target, it is imperative to understand how a complete Mac1 deletion impacts the replication and pathogenesis of different CoVs. To this end, we created recombinant bacterial artificial chromosomes (BACs) containing complete Mac1 deletions (ΔMac1) in MHV, MERS-CoV, and SARS-CoV-2. While we were unable to recover infectious virus from MHV or MERS-CoV ΔMac1 BACs, SARS-CoV-2 ΔMac1 was readily recovered from BAC transfection, indicating a stark difference in the requirement for Mac1 between different CoVs. Furthermore, SARS-CoV-2 ΔMac1 replicated at or near wild-type levels in multiple cell lines susceptible to infection. However, in a mouse model of severe infection, ΔMac1 was quickly cleared causing minimal pathology without any morbidity. ΔMac1 SARS-CoV-2 induced increased levels of interferon (IFN) and interferon-stimulated gene (ISG) expression in cell culture and mice, indicating that Mac1 blocks IFN responses which may contribute to its attenuation. ΔMac1 infection also led to a stark reduction in inflammatory monocytes and neutrophils. These results demonstrate that Mac1 only minimally impacts SARS-CoV-2 replication, unlike MHV and MERS-CoV, but is required for SARS-CoV-2 pathogenesis and is a unique antiviral drug target.
Significance: All CoVs, including SARS-CoV-2, encode for a conserved macrodomain (Mac1) that counters host ADP-ribosylation. Prior studies with SARS-CoV-1 and MHV found that Mac1 blocks IFN production and promotes CoV pathogenesis, which has prompted the development of SARS-CoV-2 Mac1 inhibitors. However, development of these compounds into antivirals requires that we understand how SARS-CoV-2 lacking Mac1 replicates and causes disease in vitro and in vivo . Here we found that SARS-CoV-2 containing a complete Mac1 deletion replicates normally in cell culture but induces an elevated IFN response, has reduced viral loads in vivo , and does not cause significant disease in mice. These results will provide a roadmap for testing Mac1 inhibitors, help identify Mac1 functions, and open additional avenues for coronavirus therapies.
Conflict of interest statement
Figures




References
-
- Beigel J. H., Tomashek K. M., Dodd L. E., Remdesivir for the Treatment of Covid-19 - Preliminary Report. Reply. N Engl J Med 383, 994 (2020). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
