This is a preprint.
Systematic visualisation of molecular QTLs reveals variant mechanisms at GWAS loci
- PMID: 37066341
- PMCID: PMC10104061
- DOI: 10.1101/2023.04.06.535816
Systematic visualisation of molecular QTLs reveals variant mechanisms at GWAS loci
Update in
-
eQTL Catalogue 2023: New datasets, X chromosome QTLs, and improved detection and visualisation of transcript-level QTLs.PLoS Genet. 2023 Sep 18;19(9):e1010932. doi: 10.1371/journal.pgen.1010932. eCollection 2023 Sep. PLoS Genet. 2023. PMID: 37721944 Free PMC article.
Abstract
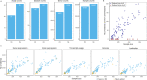
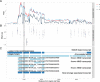
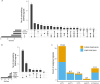
Splicing quantitative trait loci (QTLs) have been implicated as a common mechanism underlying complex trait associations. However, utilising splicing QTLs in target discovery and prioritisation has been challenging due to extensive data normalisation which often renders the direction of the genetic effect as well as its magnitude difficult to interpret. This is further complicated by the fact that strong expression QTLs often manifest as weak splicing QTLs and vice versa, making it difficult to uniquely identify the underlying molecular mechanism at each locus. We find that these ambiguities can be mitigated by visualising the association between the genotype and average RNA sequencing read coverage in the region. Here, we generate these QTL coverage plots for 1.7 million molecular QTL associations in the eQTL Catalogue identified with five quantification methods. We illustrate the utility of these QTL coverage plots by performing colocalisation between vitamin D levels in the UK Biobank and all molecular QTLs in the eQTL Catalogue. We find that while visually confirmed splicing QTLs explain just 6/53 of the colocalising signals, they are significantly less pleiotropic than eQTLs and identify a prioritised causal gene in 4/6 cases. All our association summary statistics and QTL coverage plots are freely available at https://www.ebi.ac.uk/eqtl/.
Conflict of interest statement
Competing interests J.C.U. is an employee of Illumina. N.K. is an employee of Nightingale Health. M.W. is an employee of Genomics Plc. S.G. is an employee of Verge Genomics.
Figures
References
-
- Alasoo K. 2017. wiggleplotr: Make read coverage plots from BigWig files. Bioconductor. doi:10.18129/B9.bioc.wiggleplotr - DOI
Publication types
LinkOut - more resources
Full Text Sources
