This is a preprint.
MANGEM: a web app for Multimodal Analysis of Neuronal Gene expression, Electrophysiology and Morphology
- PMID: 37066386
- PMCID: PMC10104012
- DOI: 10.1101/2023.04.03.535322
MANGEM: a web app for Multimodal Analysis of Neuronal Gene expression, Electrophysiology and Morphology
Update in
-
MANGEM: A web app for multimodal analysis of neuronal gene expression, electrophysiology, and morphology.Patterns (N Y). 2023 Sep 25;4(11):100847. doi: 10.1016/j.patter.2023.100847. eCollection 2023 Nov 10. Patterns (N Y). 2023. PMID: 38035195 Free PMC article.
Abstract
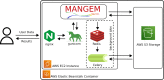
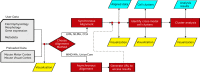
Single-cell techniques have enabled the acquisition of multi-modal data, particularly for neurons, to characterize cellular functions. Patch-seq, for example, combines patch-clamp recording, cell imaging, and single-cell RNA-seq to obtain electrophysiology, morphology, and gene expression data from a single neuron. While these multi-modal data offer potential insights into neuronal functions, they can be heterogeneous and noisy. To address this, machine-learning methods have been used to align cells from different modalities onto a low-dimensional latent space, revealing multi-modal cell clusters. However, the use of those methods can be challenging for biologists and neuroscientists without computational expertise and also requires suitable computing infrastructure for computationally expensive methods. To address these issues, we developed a cloud-based web application, MANGEM (Multimodal Analysis of Neuronal Gene expression, Electrophysiology, and Morphology) at https://ctc.waisman.wisc.edu/mangem. MANGEM provides a step-by-step accessible and user-friendly interface to machine-learning alignment methods of neuronal multi-modal data while enabling real-time visualization of characteristics of raw and aligned cells. It can be run asynchronously for large-scale data alignment, provides users with various downstream analyses of aligned cells and visualizes the analytic results such as identifying multi-modal cell clusters of cells and detecting correlated genes with electrophysiological and morphological features. We demonstrated the usage of MANGEM by aligning Patch-seq multimodal data of neuronal cells in the mouse visual cortex.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures



References
-
- Marx V. Patch-seq takes neuroscience to a multimodal place. Nat Methods. 2022. Nov;19(11):1340–4. - PubMed
-
- Institute for Genome Sciences. NeMO Archive - Home [Internet]. The Neuroscience Multi-omic Archive. [cited 2023 Mar 27]. Available from: https://nemoarchive.org/
-
- DANDI Archive [Internet]. [cited 2023 Mar 27]. Available from: https://dandiarchive.org/
-
- Efremova M, Teichmann SA. Computational methods for single-cell omics across modalities. Nat Methods. 2020. Jan;17(1):14–7. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
