Hepatocyte SREBP signaling mediates clock communication within the liver
- PMID: 37066875
- PMCID: PMC10104893
- DOI: 10.1172/JCI163018
Hepatocyte SREBP signaling mediates clock communication within the liver
Abstract
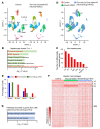
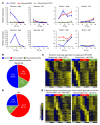
Rhythmic intraorgan communication coordinates environmental signals and the cell-intrinsic clock to maintain organ homeostasis. Hepatocyte-specific KO of core components of the molecular clock Rev-erbα and -β (Reverb-hDKO) alters cholesterol and lipid metabolism in hepatocytes as well as rhythmic gene expression in nonparenchymal cells (NPCs) of the liver. Here, we report that in fatty liver caused by diet-induced obesity (DIO), hepatocyte SREBP cleavage-activating protein (SCAP) was required for Reverb-hDKO-induced diurnal rhythmic remodeling and epigenomic reprogramming in liver macrophages (LMs). Integrative analyses of isolated hepatocytes and LMs revealed that SCAP-dependent lipidomic changes in REV-ERB-depleted hepatocytes led to the enhancement of LM metabolic rhythms. Hepatocytic loss of REV-ERBα and β (REV-ERBs) also attenuated LM rhythms via SCAP-independent polypeptide secretion. These results shed light on the signaling mechanisms by which hepatocytes regulate diurnal rhythms in NPCs in fatty liver disease caused by DIO.
Keywords: Epigenetics; Hepatology; Homeostasis; Metabolism.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

