High Genetic Diversity of Carbapenem-Resistant Acinetobacter baumannii Isolates Recovered in Nigerian Hospitals in 2016 to 2020
- PMID: 37067411
- PMCID: PMC10286719
- DOI: 10.1128/msphere.00098-23
High Genetic Diversity of Carbapenem-Resistant Acinetobacter baumannii Isolates Recovered in Nigerian Hospitals in 2016 to 2020
Erratum in
-
Correction for Odih et al., "High Genetic Diversity of Carbapenem-Resistant Acinetobacter baumannii Isolates Recovered in Nigerian Hospitals in 2016 to 2020".mSphere. 2024 Jan 30;9(1):e0065923. doi: 10.1128/msphere.00659-23. Epub 2023 Dec 21. mSphere. 2024. PMID: 38126760 Free PMC article. No abstract available.
Abstract
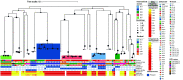
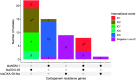
Acinetobacter baumannii causes difficult-to-treat infections mostly among immunocompromised patients. Clinically relevant A. baumannii lineages and their carbapenem resistance mechanisms are sparsely described in Nigeria. This study aimed to characterize the diversity and genetic mechanisms of carbapenem resistance among A. baumannii strains isolated from hospitals in southwestern Nigeria. We sequenced the genomes of all A. baumannii isolates submitted to Nigeria's antimicrobial resistance surveillance reference laboratory between 2016 and 2020 on an Illumina platform and performed in silico genomic characterization. Selected strains were sequenced using the Oxford Nanopore technology to characterize the genetic context of carbapenem resistance genes. The 86 A. baumannii isolates were phylogenetically diverse and belonged to 35 distinct Oxford sequence types (oxfSTs), 16 of which were novel, and 28 Institut Pasteur STs (pasSTs). Thirty-eight (44.2%) isolates belonged to none of the known international clones (ICs). Over 50% of the isolates were phenotypically resistant to 10 of 12 tested antimicrobials. The majority (n = 54) of the isolates were carbapenem resistant, particularly the IC7 (pasST25; 100%) and IC9 (pasST85; >91.7%) strains. blaOXA-23 (34.9%) and blaNDM-1 (27.9%) were the most common carbapenem resistance genes detected. All blaOXA-23 genes were carried on Tn2006 or Tn2006-like transposons. Our findings suggest that a 10-kb Tn125 composite transposon is the primary means of blaNDM-1 dissemination. Our findings highlight an increase in blaNDM-1 prevalence and the widespread transposon-facilitated dissemination of carbapenemase genes in diverse A. baumannii lineages in southwestern Nigeria. We make the case for improving surveillance of these pathogens in Nigeria and other understudied settings. IMPORTANCE Acinetobacter baumannii bacteria are increasingly clinically relevant due to their propensity to harbor genes conferring resistance to multiple antimicrobials, as well as their ability to persist and disseminate in hospital environments and cause difficult-to-treat nosocomial infections. Little is known about the molecular epidemiology and antimicrobial resistance profiles of these organisms in Nigeria, largely due to limited capacity for their isolation, identification, and antimicrobial susceptibility testing. Our study characterized the diversity and antimicrobial resistance profiles of clinical A. baumannii in southwestern Nigeria using whole-genome sequencing. We also identified the key genetic elements facilitating the dissemination of carbapenem resistance genes within this species. This study provides key insights into the clinical burden and population dynamics of A. baumannii in hospitals in Nigeria and highlights the importance of routine whole-genome sequencing-based surveillance of this and other previously understudied pathogens in Nigeria and other similar settings.
Keywords: Acinetobacter baumannii; antimicrobial resistance; blaNDM-1; blaOXA-23; carbapenem resistance; genomics; surveillance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
