Antimicrobial-Resistant Enterobacterales Recovered from the Environment of Two Zoological Institutions Include Enterobacter cloacae Complex ST171 Producing KPC-4 Carbapenemase
- PMID: 37067417
- PMCID: PMC10231243
- DOI: 10.1128/aem.00257-23
Antimicrobial-Resistant Enterobacterales Recovered from the Environment of Two Zoological Institutions Include Enterobacter cloacae Complex ST171 Producing KPC-4 Carbapenemase
Abstract
Environmental surfaces can serve as reservoirs for pathogens and antimicrobial-resistant (AMR) bacteria in healthcare settings. Although active surveillance programs are used in veterinary and human healthcare, unconventional settings like zoological facilities are often overlooked, even though antimicrobials are used to maintain the health of their animal collections. Here, we used electrostatic cloths to conduct active environmental surveillance over a 2-year period at two zoological institutions to determine contamination prevalence of human-only and mixed animal-human touch environments with AMR bacteria. We recovered Enterobacterales isolates that expressed quinolone resistance, an AmpC-like phenotype, and an extended-spectrum β-lactamase phenotype from 144 (39%), 141 (38.2%), and 72 (19.5%) of the environmental samples, respectively. The zoological institutions, areas and exhibits within the zoological facility, and sampling surface type affected the odds of recovering AMR bacteria from the environment. Three carbapenemase-producing Enterobacter cloacae complex ST171 isolates recovered from one zoological facility harbored an IncH12 plasmid with a Tn4401b-KPC-4 transposon conferring multidrug resistance. One isolate maintained three tandem repeats of a Tn4401b-KPC-4 element on an IncHI2 plasmid, although this isolate was susceptible to the four carbapenem drugs tested. These three isolates and their IncH12 plasmids shared significant genomic similarity with two E. cloacae complex isolates recovered from canine patients at a regional veterinary hospital during year 2 of this study. Our results indicated that surface environments at zoological institutions can serve as reservoirs for AMR bacteria and their genes and have implications for animal and public health. IMPORTANCE Environmental surfaces can be a source of antimicrobial-resistant (AMR) bacteria that pose a risk to human and animal health. Zoological institutions are unique environments where exotic animals, staff, and visitors intermingle and antimicrobials are used to maintain animal health. However, zoological environments are often overlooked as reservoirs of AMR bacteria. Here, we show that zoological environments can serve as reservoirs of fluoroquinolone-resistant and extended-spectrum cephalosporin-resistant bacteria. In addition, we isolated three carbapenemase-producing Enterobacter cloacae complex strains carrying blaKPC-4, including one with a unique, tandem triplicate of the Tn4401b-KPC-4 element. Comparative whole genomics of these isolates with two E. cloacae complex isolates from patients at a regional veterinary hospital highlighted the possibility of local KPC-4 spread between animal environments. Our results suggest that environments at zoological institutions serve as reservoirs for AMR bacteria and pose a hypothetical One Health risk to the public, staff, and the wild animal populations in captivity.
Keywords: carbapenemase-producing Enterobacterales; cephalosporin-resistant; fluoroquinolone-resistant; veterinary epidemiology; zoological environments.
Conflict of interest statement
The authors declare no conflict of interest.
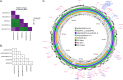
Figures


References
-
- Lockhart SR, Abramson MA, Beekmann SE, Gallagher G, Riedel S, Diekema DJ, Quinn JP, Doern GV. 2007. Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J Clin Microbiol 45:3352–3359. doi:10.1128/JCM.01284-07. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

