Adaptive Regulation of Stopover Refueling during Bird Migration: Insights from Whole Blood Transcriptomics
- PMID: 37067540
- PMCID: PMC10139441
- DOI: 10.1093/gbe/evad061
Adaptive Regulation of Stopover Refueling during Bird Migration: Insights from Whole Blood Transcriptomics
Abstract
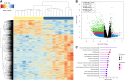
Migration is one of the most energy-demanding tasks in avian life cycle. Many birds might not have sufficient fuel stores to cover long distances, so they must stop to rest and refuel at stopover sites, especially after the crossing of large ecological barriers. There, birds undergo several behavioral, morphological, and physiological trait adjustments to recover from and prepare for their journey; however, regulation of such processes at the molecular level remains largely unknown. In this study, we used transcriptomic information from the whole blood of migrating garden warblers (Sylvia borin) to identify key regulatory pathways related to adaptations for migration. Birds were temporarily caged during spring migration stopover and then sampled twice at different refueling states (lean vs. fat), reflecting different migratory stages (stopover arrival vs. departure) after the crossing of an extended ecological barrier. Our results show that top expressed genes during migration are involved in important pathways regarding adaptations to migration at high altitudes such as increase of aerobic capacity and angiogenesis. Gene expression profiles largely reflected the two experimental conditions with several enzymes involved in different aspects of metabolic activity being differentially expressed between states providing several candidate genes for future functional studies. Additionally, we identified several hub genes, upregulated in lean birds that could be involved in the extraordinary phenotypic flexibility in organ mass displayed by avian migrants. Finally, our approach provides novel evidence that regulation of water homeostasis may represent a significant adaptive mechanism, allowing birds to conserve water during long-distance flight, mainly through protein catabolism.
Keywords: RNA-seq; candidate genes; differential gene expression; metabolism; phenotypic plasticity; water homeostasis.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


References
-
- Åkesson S, Hedenström A. 2007. How migrants get there: migratory performance and orientation. BioScience 57:123–133.
-
- Alerstam T. 2011. Optimal bird migration revisited. J Ornithol. 152:5–23.
-
- Alerstam T, Lindström Å. 1990. Optimal bird migration: the relative importance of time, energy, and safety. In: Gwinner E, editors. Bird migration. Berlin: Springer. p. 331–351.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

