A frame-shift mutation in COMTD1 is associated with impaired pheomelanin pigmentation in chicken
- PMID: 37068079
- PMCID: PMC10138217
- DOI: 10.1371/journal.pgen.1010724
A frame-shift mutation in COMTD1 is associated with impaired pheomelanin pigmentation in chicken
Abstract
The biochemical pathway regulating the synthesis of yellow/red pheomelanin is less well characterized than the synthesis of black/brown eumelanin. Inhibitor of gold (IG phenotype) is a plumage colour variant in chicken that provides an opportunity to further explore this pathway since the recessive allele (IG) at this locus is associated with a defect in the production of pheomelanin. IG/IG homozygotes display a marked dilution of red pheomelanin pigmentation, whilst black pigmentation (eumelanin) is only slightly affected. Here we show that a 2-base pair insertion (frame-shift mutation) in the 5th exon of the Catechol-O-methyltransferase containing domain 1 gene (COMTD1), expected to cause a complete or partial loss-of-function of the COMTD1 enzyme, shows complete concordance with the IG phenotype within and across breeds. We show that the COMTD1 protein is localized to mitochondria in pigment cells. Knockout of Comtd1 in a mouse melanocytic cell line results in a reduction in pheomelanin metabolites and significant alterations in metabolites of glutamate/glutathione, riboflavin, and the tricarboxylic acid cycle. Furthermore, COMTD1 overexpression enhanced cellular proliferation following chemical-induced transfection, a potential inducer of oxidative stress. These observations suggest that COMTD1 plays a protective role for melanocytes against oxidative stress and that this supports their ability to produce pheomelanin.
Copyright: © 2023 Bi et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
No competing interest.
Figures




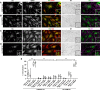


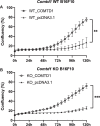
References
-
- Zadlo A, Szewczyk G, Sarna M, Camenisch TG, Sidabras JW, Ito S, et al.. Photobleaching of pheomelanin increases its phototoxic potential: Physicochemical studies of synthetic pheomelanin subjected to aerobic photolysis. Pigment Cell Melanoma Res. 2019;32:359–72. doi: 10.1111/pcmr.12752 . - DOI - PubMed
-
- Chedekel MR, Agin PP, Sayre RM. Photochemistry of pheomelanin: action spectrum for superoxide production. Photochem Photobiol. 1980;31:553–5. doi: 10.1111/j.1751-1097.1980.tb03745.x - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

