A comparative 'omics' approach for prediction of candidate Strongyloides stercoralis diagnostic coproantigens
- PMID: 37068106
- PMCID: PMC10138266
- DOI: 10.1371/journal.pntd.0010777
A comparative 'omics' approach for prediction of candidate Strongyloides stercoralis diagnostic coproantigens
Abstract
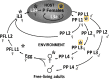
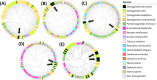
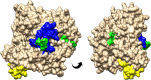
Human infection with the intestinal nematode Strongyloides stercoralis is persistent unless effectively treated, and potentially fatal in immunosuppressed individuals. Epidemiological data are lacking, partially due to inadequate diagnosis. A rapid antigen detection test is a priority for population surveillance, validating cure after treatment, and for screening prior to immunosuppression. We used a targeted analysis of open access 'omics' data sets and used online predictors to identify S. stercoralis proteins that are predicted to be present in infected stool, Strongyloides-specific, and antigenic. Transcriptomic data from gut and non-gut dwelling life cycle stages of S. stercoralis revealed 328 proteins that are differentially expressed. Strongyloides ratti proteomic data for excreted and secreted (E/S) proteins were matched to S. stercoralis, giving 1,057 orthologues. Five parasitism-associated protein families (SCP/TAPS, prolyl oligopeptidase, transthyretin-like, aspartic peptidase, acetylcholinesterase) were compared phylogenetically between S. stercoralis and outgroups, and proteins with least homology to the outgroups were selected. Proteins that overlapped between the transcriptomic and proteomic datasets were analysed by multiple sequence alignment, epitope prediction and 3D structure modelling to reveal S. stercoralis candidate peptide/protein coproantigens. We describe 22 candidates from seven genes, across all five protein families for further investigation as potential S. stercoralis diagnostic coproantigens, identified using open access data and freely-available protein analysis tools. This powerful approach can be applied to many parasitic infections with 'omic' data to accelerate development of specific diagnostic assays for laboratory or point-of-care field application.
Copyright: © 2023 Marlais et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Buonfrate D, Baldissera M, Abrescia F, Bassetti M, Caramaschi G, Giobbia M, et al.. Epidemiology of Strongyloides stercoralis in northern Italy: results of a multicentre case-control study, February 2013 to July 2014. Euro Surveill. 2016; 21(31). doi: 10.2807/1560-7917.ES.2016.21.31.30310 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

