The viral packaging motor potentiates Kaposi's sarcoma-associated herpesvirus gene expression late in infection
- PMID: 37068108
- PMCID: PMC10138851
- DOI: 10.1371/journal.ppat.1011163
The viral packaging motor potentiates Kaposi's sarcoma-associated herpesvirus gene expression late in infection
Abstract
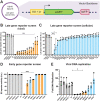
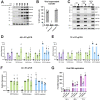
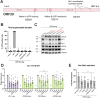
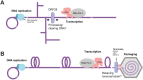
β- and γ-herpesviruses transcribe their late genes in a manner distinct from host transcription. This process is directed by a complex of viral transcriptional activator proteins that hijack cellular RNA polymerase II and an unknown set of additional factors. We employed proximity labeling coupled with mass spectrometry, followed by CRISPR and siRNA screening to identify proteins functionally associated with the Kaposi's sarcoma-associated herpesvirus (KSHV) late gene transcriptional complex. These data revealed that the catalytic subunit of the viral DNA packaging motor, ORF29, is both dynamically associated with the viral transcriptional activator complex and potentiates gene expression late in infection. Through genetic mutation and deletion of ORF29, we establish that its catalytic activity potentiates viral transcription and is required for robust accumulation of essential late proteins during infection. Thus, we propose an expanded role for ORF29 that encompasses its established function in viral packaging and its newly discovered contributions to viral transcription and late gene expression in KSHV.
Copyright: © 2023 McCollum et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Kaposi's Sarcoma-Associated Herpesvirus Hijacks RNA Polymerase II To Create a Viral Transcriptional Factory.J Virol. 2017 May 12;91(11):e02491-16. doi: 10.1128/JVI.02491-16. Print 2017 Jun 1. J Virol. 2017. PMID: 28331082 Free PMC article.
-
Conserved CxnC Motifs in Kaposi's Sarcoma-Associated Herpesvirus ORF66 Are Required for Viral Late Gene Expression and Are Essential for Its Interaction with ORF34.J Virol. 2020 Jan 6;94(2):e01299-19. doi: 10.1128/JVI.01299-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31578296 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus ORF67.5 Functions as a Component of the Terminase Complex.J Virol. 2023 Jun 29;97(6):e0047523. doi: 10.1128/jvi.00475-23. Epub 2023 Jun 5. J Virol. 2023. PMID: 37272800 Free PMC article.
-
[Replication Machinery of Kaposi's Sarcoma-associated Herpesvirus and Drug Discovery Research].Yakugaku Zasshi. 2019;139(1):69-73. doi: 10.1248/yakushi.18-00164-2. Yakugaku Zasshi. 2019. PMID: 30606932 Review. Japanese.
-
Posttranscriptional gene regulation in Kaposi's sarcoma-associated herpesvirus.Adv Appl Microbiol. 2009;68:241-61. doi: 10.1016/S0065-2164(09)01206-4. Adv Appl Microbiol. 2009. PMID: 19426857 Review.
Cited by
-
The elevated expression of ORF75, a KSHV lytic gene, in Kaposi sarcoma lesions is driven by a GC-rich DNA cis element in its promoter region.PLoS Pathog. 2025 Mar 17;21(3):e1012984. doi: 10.1371/journal.ppat.1012984. eCollection 2025 Mar. PLoS Pathog. 2025. PMID: 40096169 Free PMC article.
-
Enhancers and genome conformation provide complex transcriptional control of a herpesviral gene.Mol Syst Biol. 2025 Jan;21(1):30-58. doi: 10.1038/s44320-024-00075-0. Epub 2024 Nov 19. Mol Syst Biol. 2025. PMID: 39562742 Free PMC article.
-
General Maintenance and Reactivation of iSLK Cell Lines.Bio Protoc. 2025 Jun 5;15(11):e5334. doi: 10.21769/BioProtoc.5334. eCollection 2025 Jun 5. Bio Protoc. 2025. PMID: 40511416 Free PMC article.
-
From enhancers to genome conformation: complex transcriptional control underlies expression of a single herpesviral gene.bioRxiv [Preprint]. 2024 Jun 16:2023.07.08.548212. doi: 10.1101/2023.07.08.548212. bioRxiv. 2024. Update in: Mol Syst Biol. 2025 Jan;21(1):30-58. doi: 10.1038/s44320-024-00075-0. PMID: 37461644 Free PMC article. Updated. Preprint.
References
-
- Nandakumar D, Glaunsinger B. An integrative approach identifies direct targets of the late viral transcription complex and an expanded promoter recognition motif in Kaposi’s sarcoma-associated herpesvirus. PLoS Pathog. 2019;15(5):e1007774. Epub 20190516. doi: 10.1371/journal.ppat.1007774 ; PubMed Central PMCID: PMC6541308. - DOI - PMC - PubMed
-
- Li M, Hu Q, Collins G, Parida M, Ball CB, Price DH, et al.. Cytomegalovirus late transcription factor target sequence diversity orchestrates viral early to late transcription. PLoS Pathog. 2021;17(8):e1009796. Epub 20210802. doi: 10.1371/journal.ppat.1009796 ; PubMed Central PMCID: PMC8360532. - DOI - PMC - PubMed
-
- Aubry V, Mure F, Mariame B, Deschamps T, Wyrwicz LS, Manet E, et al.. Epstein-Barr virus late gene transcription depends on the assembly of a virus-specific preinitiation complex. J Virol. 2014;88(21):12825–38. Epub 20140827. doi: 10.1128/JVI.02139-14 ; PubMed Central PMCID: PMC4248913. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

