Computational drug discovery for castration-resistant prostate cancers through in vitro drug response modeling
- PMID: 37068243
- PMCID: PMC10151558
- DOI: 10.1073/pnas.2218522120
Computational drug discovery for castration-resistant prostate cancers through in vitro drug response modeling
Abstract
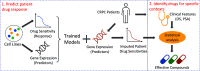
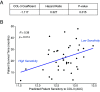
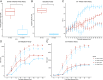
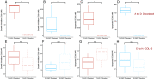
Prostate cancer (PC) is the most frequently diagnosed malignancy and a leading cause of cancer deaths in US men. Many PC cases metastasize and develop resistance to systemic hormonal therapy, a stage known as castration-resistant prostate cancer (CRPC). Therefore, there is an urgent need to develop effective therapeutic strategies for CRPC. Traditional drug discovery pipelines require significant time and capital input, which highlights a need for novel methods to evaluate the repositioning potential of existing drugs. Here, we present a computational framework to predict drug sensitivities of clinical CRPC tumors to various existing compounds and identify treatment options with high potential for clinical impact. We applied this method to a CRPC patient cohort and nominated drugs to combat resistance to hormonal therapies including abiraterone and enzalutamide. The utility of this method was demonstrated by nomination of multiple drugs that are currently undergoing clinical trials for CRPC. Additionally, this method identified the tetracycline derivative COL-3, for which we validated higher efficacy in an isogenic cell line model of enzalutamide-resistant vs. enzalutamide-sensitive CRPC. In enzalutamide-resistant CRPC cells, COL-3 displayed higher activity for inhibiting cell growth and migration, and for inducing G1-phase cell cycle arrest and apoptosis. Collectively, these findings demonstrate the utility of a computational framework for independent validation of drugs being tested in CRPC clinical trials, and for nominating drugs with enhanced biological activity in models of enzalutamide-resistant CRPC. The efficiency of this method relative to traditional drug development approaches indicates a high potential for accelerating drug development for CRPC.
Keywords: castration-resistant prostate cancer; drug discovery; drug repurpose; drug response prediction; enzalutamide.
Conflict of interest statement
The authors declare no competing interest.
Figures


Comment in
-
Uro-Science.J Urol. 2024 Jan;211(1):194-195. doi: 10.1097/JU.0000000000003710. Epub 2023 Oct 20. J Urol. 2024. PMID: 37861082 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

