Anionic G•U pairs in bacterial ribosomal rRNAs
- PMID: 37068913
- PMCID: PMC10275268
- DOI: 10.1261/rna.079583.123
Anionic G•U pairs in bacterial ribosomal rRNAs
Abstract
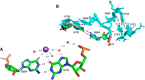
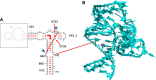
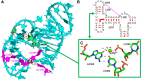
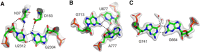
Wobble GU pairs (or G•U) occur frequently within double-stranded RNA helices interspersed between standard G=C and A-U Watson-Crick pairs. Another type of G•U pair interacting via their Watson-Crick edges has been observed in the A site of ribosome structures between a modified U34 in the tRNA anticodon triplet and G + 3 in the mRNA. In such pairs, the electronic structure of the U is changed with a negative charge on N3(U), resulting in two H-bonds between N1(G)…O4(U) and N2(G)…N3(U). Here, we report that such pairs occur in other highly conserved positions in ribosomal RNAs of bacteria in the absence of U modification. An anionic cis Watson-Crick G•G pair is also observed and well conserved in the small subunit. These pairs are observed in tightly folded regions.
Keywords: G•U pair; anionic; anticodon; codon; mRNA; rRNA; tRNA.
© 2023 Westhof et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures




Similar articles
-
The multiple flavors of GoU pairs in RNA.J Mol Recognit. 2019 Aug;32(8):e2782. doi: 10.1002/jmr.2782. Epub 2019 Apr 29. J Mol Recognit. 2019. PMID: 31033092 Free PMC article. Review.
-
Tautomeric G•U pairs within the molecular ribosomal grip and fidelity of decoding in bacteria.Nucleic Acids Res. 2018 Aug 21;46(14):7425-7435. doi: 10.1093/nar/gky547. Nucleic Acids Res. 2018. PMID: 29931292 Free PMC article.
-
Stacking of Crick Wobble pair and Watson-Crick pair: stability rules of G-U pairs at ends of helical stems in tRNAs and the relation to codon-anticodon Wobble interaction.Nucleic Acids Res. 1978 Nov;5(11):4451-61. doi: 10.1093/nar/5.11.4451. Nucleic Acids Res. 1978. PMID: 724522 Free PMC article.
-
New structural insights into the decoding mechanism: translation infidelity via a G·U pair with Watson-Crick geometry.FEBS Lett. 2013 Jun 27;587(13):1848-57. doi: 10.1016/j.febslet.2013.05.009. Epub 2013 May 23. FEBS Lett. 2013. PMID: 23707250 Review.
-
The influence of anticodon-codon interactions and modified bases on codon usage bias in bacteria.Mol Biol Evol. 2010 Sep;27(9):2129-40. doi: 10.1093/molbev/msq102. Epub 2010 Apr 19. Mol Biol Evol. 2010. PMID: 20403966
Cited by
-
Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.Nat Struct Mol Biol. 2024 May;31(5):817-825. doi: 10.1038/s41594-024-01238-1. Epub 2024 Mar 27. Nat Struct Mol Biol. 2024. PMID: 38538915
-
Identification and characterization of shifted G•U wobble pairs resulting from alternative protonation of RNA.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf575. doi: 10.1093/nar/gkaf575. Nucleic Acids Res. 2025. PMID: 40694854 Free PMC article.
-
NMR measurements of transient low-populated tautomeric and anionic Watson-Crick-like G·T/U in RNA:DNA hybrids: implications for the fidelity of transcription and CRISPR/Cas9 gene editing.Nucleic Acids Res. 2024 Mar 21;52(5):2672-2685. doi: 10.1093/nar/gkae027. Nucleic Acids Res. 2024. PMID: 38281263 Free PMC article.
-
The role of ribosomal protein networks in ribosome dynamics.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1308. doi: 10.1093/nar/gkae1308. Nucleic Acids Res. 2025. PMID: 39788545 Free PMC article.
-
A general strategy for engineering GU base pairs to facilitate RNA crystallization.Nucleic Acids Res. 2025 Jan 24;53(3):gkae1218. doi: 10.1093/nar/gkae1218. Nucleic Acids Res. 2025. PMID: 39721592 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
