ARRDC5 expression is conserved in mammalian testes and required for normal sperm morphogenesis
- PMID: 37069147
- PMCID: PMC10110545
- DOI: 10.1038/s41467-023-37735-y
ARRDC5 expression is conserved in mammalian testes and required for normal sperm morphogenesis
Abstract
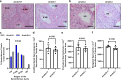
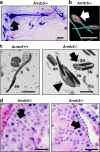
In sexual reproduction, sperm contribute half the genomic material required for creation of offspring yet core molecular mechanisms essential for their formation are undefined. Here, the α-arrestin molecule arrestin-domain containing 5 (ARRDC5) is identified as an essential regulator of mammalian spermatogenesis. Multispecies testicular tissue transcriptome profiling indicates that expression of Arrdc5 is testis enriched, if not specific, in mice, pigs, cattle, and humans. Knockout of Arrdc5 in mice leads to male specific sterility due to production of low numbers of sperm that are immotile and malformed. Spermiogenesis, the final phase of spermatogenesis when round spermatids transform to spermatozoa, is defective in testes of Arrdc5 deficient mice. Also, epididymal sperm in Arrdc5 knockouts are unable to capacitate and fertilize oocytes. These findings establish ARRDC5 as an essential regulator of mammalian spermatogenesis. Considering the role of arrestin molecules as modulators of cellular signaling and ubiquitination, ARRDC5 is a potential male contraceptive target.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Russell, L. D., Ettlin, R. A., Hikim, A. P. S & Cless, E. D. Histological and Histopathological Evaluation of the Testis 1st edn (Cache River Press, 1990).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

