DRAG in situ barcoding reveals an increased number of HSPCs contributing to myelopoiesis with age
- PMID: 37069150
- PMCID: PMC10110593
- DOI: 10.1038/s41467-023-37167-8
DRAG in situ barcoding reveals an increased number of HSPCs contributing to myelopoiesis with age
Abstract
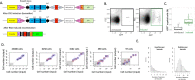
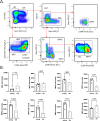
Ageing is associated with changes in the cellular composition of the immune system. During ageing, hematopoietic stem and progenitor cells (HSPCs) that produce immune cells are thought to decline in their regenerative capacity. However, HSPC function has been mostly assessed using transplantation assays, and it remains unclear how HSPCs age in the native bone marrow niche. To address this issue, we present an in situ single cell lineage tracing technology to quantify the clonal composition and cell production of single cells in their native niche. Our results demonstrate that a pool of HSPCs with unequal output maintains myelopoiesis through overlapping waves of cell production throughout adult life. During ageing, the increased frequency of myeloid cells is explained by greater numbers of HSPCs contributing to myelopoiesis rather than the increased myeloid output of individual HSPCs. Strikingly, the myeloid output of HSPCs remains constant over time despite accumulating significant transcriptomic changes throughout adulthood. Together, these results show that, unlike emergency myelopoiesis post-transplantation, aged HSPCs in their native microenvironment do not functionally decline in their regenerative capacity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Different Human Immune Lineage Compositions Are Generated in Non-Conditioned NBSGW Mice Depending on HSPC Source.Front Immunol. 2020 Oct 19;11:573406. doi: 10.3389/fimmu.2020.573406. eCollection 2020. Front Immunol. 2020. PMID: 33193358 Free PMC article.
-
Breast cancer remotely imposes a myeloid bias on haematopoietic stem cells by reprogramming the bone marrow niche.Nat Cell Biol. 2023 Dec;25(12):1736-1745. doi: 10.1038/s41556-023-01291-w. Epub 2023 Nov 30. Nat Cell Biol. 2023. PMID: 38036749
-
Diabetes-Associated Myelopoiesis Drives Stem Cell Mobilopathy Through an OSM-p66Shc Signaling Pathway.Diabetes. 2019 Jun;68(6):1303-1314. doi: 10.2337/db19-0080. Epub 2019 Apr 1. Diabetes. 2019. PMID: 30936144
-
Emergency myelopoiesis in solid cancers.Br J Haematol. 2024 Sep;205(3):798-811. doi: 10.1111/bjh.19656. Epub 2024 Jul 23. Br J Haematol. 2024. PMID: 39044285 Free PMC article. Review.
-
Roles of Hematopoietic Stem and Progenitor Cells in Ischemic Cardiovascular Disease.Curr Stem Cell Res Ther. 2021;16(5):589-598. doi: 10.2174/1574888X15666200130091858. Curr Stem Cell Res Ther. 2021. PMID: 32000654 Review.
Cited by
-
Extracting, filtering and simulating cellular barcodes using CellBarcode tools.Nat Comput Sci. 2024 Feb;4(2):128-143. doi: 10.1038/s43588-024-00595-7. Epub 2024 Feb 19. Nat Comput Sci. 2024. PMID: 38374363 Free PMC article.
-
Somatic epimutations enable single-cell lineage tracing in native hematopoiesis across the murine and human lifespan.bioRxiv [Preprint]. 2024 Apr 1:2024.04.01.587514. doi: 10.1101/2024.04.01.587514. bioRxiv. 2024. Update in: Nature. 2025 Jul;643(8071):478-487. doi: 10.1038/s41586-025-09041-8. PMID: 38617287 Free PMC article. Updated. Preprint.
-
Hematopoietic stem cell a reservoir of innate immune memory.Front Immunol. 2024 Dec 10;15:1491729. doi: 10.3389/fimmu.2024.1491729. eCollection 2024. Front Immunol. 2024. PMID: 39720722 Free PMC article. Review.
-
Hematopoietic Stem Cell Fates and the Cellular Hierarchy of Mammalian Hematopoiesis: from Transplantation Models to New Insights from in Situ Analyses.Stem Cell Rev Rep. 2025 Jan;21(1):28-44. doi: 10.1007/s12015-024-10782-8. Epub 2024 Sep 2. Stem Cell Rev Rep. 2025. PMID: 39222178 Review.
-
iVAE: an interpretable representation learning framework enhances clustering performance for single-cell data.BMC Biol. 2025 Jul 15;23(1):213. doi: 10.1186/s12915-025-02315-7. BMC Biol. 2025. PMID: 40660220 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

