Chromosome-level genome assembly and population genomic resource to accelerate orphan crop lablab breeding
- PMID: 37069152
- PMCID: PMC10110558
- DOI: 10.1038/s41467-023-37489-7
Chromosome-level genome assembly and population genomic resource to accelerate orphan crop lablab breeding
Abstract
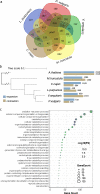
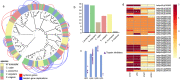
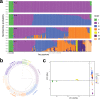
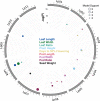
Under-utilised orphan crops hold the key to diversified and climate-resilient food systems. Here, we report on orphan crop genomics using the case of Lablab purpureus (L.) Sweet (lablab) - a legume native to Africa and cultivated throughout the tropics for food and forage. Our Africa-led plant genome collaboration produces a high-quality chromosome-scale assembly of the lablab genome. Our assembly highlights the genome organisation of the trypsin inhibitor genes - an important anti-nutritional factor in lablab. We also re-sequence cultivated and wild lablab accessions from Africa confirming two domestication events. Finally, we examine the genetic and phenotypic diversity in a comprehensive lablab germplasm collection and identify genomic loci underlying variation of important agronomic traits in lablab. The genomic data generated here provide a valuable resource for lablab improvement. Our inclusive collaborative approach also presents an example that can be explored by other researchers sequencing indigenous crops, particularly from low and middle-income countries (LMIC).
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- FAO. Faostat: FAO Statistical Databases. (Food & Agriculture Organization of the United Nations (FAO), 2000).
-
- The war in Ukraine is exposing gaps in the world’s food-systems research. Nature604, 217–218 (2022). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

