Genetically encoded barcodes for correlative volume electron microscopy
- PMID: 37069313
- PMCID: PMC10713455
- DOI: 10.1038/s41587-023-01713-y
Genetically encoded barcodes for correlative volume electron microscopy
Abstract
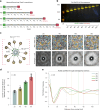
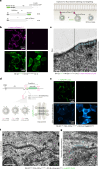
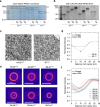
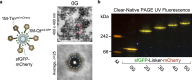
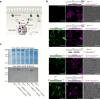
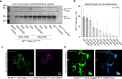
While genetically encoded reporters are common for fluorescence microscopy, equivalent multiplexable gene reporters for electron microscopy (EM) are still scarce. Here, by installing a variable number of fixation-stable metal-interacting moieties in the lumen of encapsulin nanocompartments of different sizes, we developed a suite of spherically symmetric and concentric barcodes (EMcapsulins) that are readable by standard EM techniques. Six classes of EMcapsulins could be automatically segmented and differentiated. The coding capacity was further increased by arranging several EMcapsulins into distinct patterns via a set of rigid spacers of variable length. Fluorescent EMcapsulins were expressed to monitor subcellular structures in light and EM. Neuronal expression in Drosophila and mouse brains enabled the automatic identification of genetically defined cells in EM. EMcapsulins are compatible with transmission EM, scanning EM and focused ion beam scanning EM. The expandable palette of genetically controlled EM-readable barcodes can augment anatomical EM images with multiplexed gene expression maps.
© 2023. The Author(s).
Conflict of interest statement
G.G.W., F.S., A.P. and O.B. have filed a patent application related to this paper. J.K. discloses financial interests in ariadne.ai. All other authors declare no competing interests.
Figures







References
-
- Heinrich, L. et al. Whole-cell organelle segmentation in volume electron microscopy. Nature599, 141–146 (2021). - PubMed
-
- Willingham, M. C. & Rutherford, A. V. The use of osmium-thiocarbohydrazide-osmium (OTO) and ferrocyanide-reduced osmium methods to enhance membrane contrast and preservation in cultured cells. J. Histochem. Cytochem.32, 455–460 (1984). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

