Genomics of perivascular space burden unravels early mechanisms of cerebral small vessel disease
- PMID: 37069360
- PMCID: PMC10115645
- DOI: 10.1038/s41591-023-02268-w
Genomics of perivascular space burden unravels early mechanisms of cerebral small vessel disease
Abstract
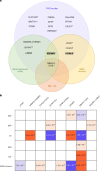
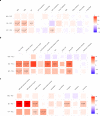
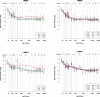
Perivascular space (PVS) burden is an emerging, poorly understood, magnetic resonance imaging marker of cerebral small vessel disease, a leading cause of stroke and dementia. Genome-wide association studies in up to 40,095 participants (18 population-based cohorts, 66.3 ± 8.6 yr, 96.9% European ancestry) revealed 24 genome-wide significant PVS risk loci, mainly in the white matter. These were associated with white matter PVS already in young adults (N = 1,748; 22.1 ± 2.3 yr) and were enriched in early-onset leukodystrophy genes and genes expressed in fetal brain endothelial cells, suggesting early-life mechanisms. In total, 53% of white matter PVS risk loci showed nominally significant associations (27% after multiple-testing correction) in a Japanese population-based cohort (N = 2,862; 68.3 ± 5.3 yr). Mendelian randomization supported causal associations of high blood pressure with basal ganglia and hippocampal PVS, and of basal ganglia PVS and hippocampal PVS with stroke, accounting for blood pressure. Our findings provide insight into the biology of PVS and cerebral small vessel disease, pointing to pathways involving extracellular matrix, membrane transport and developmental processes, and the potential for genetically informed prioritization of drug targets.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Comment in
-
Genomics of perivascular space burden across the lifespan and across ancestries.Nat Med. 2023 Apr;29(4):799-800. doi: 10.1038/s41591-023-02269-9. Nat Med. 2023. PMID: 37045999 No abstract available.
References
Publication types
MeSH terms
Grants and funding
- R01 AG054076/AG/NIA NIH HHS/United States
- R01 AG059725/AG/NIA NIH HHS/United States
- R01 AG049607/AG/NIA NIH HHS/United States
- MR/K026992/1/MRC_/Medical Research Council/United Kingdom
- N01 HC025195/HL/NHLBI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- R01 NS029993/NS/NINDS NIH HHS/United States
- U01 AG052409/AG/NIA NIH HHS/United States
- 75N92019D00031/HL/NHLBI NIH HHS/United States
- R01 AG025941/AG/NIA NIH HHS/United States
- R01 NS017950/NS/NINDS NIH HHS/United States
- MR/J006971/1/MRC_/Medical Research Council/United Kingdom
- R56 NS029993/NS/NINDS NIH HHS/United States
- BB/F019394/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- RF1 AG059421/AG/NIA NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- N02 HL064278/HL/NHLBI NIH HHS/United States
- R37 NS029993/NS/NINDS NIH HHS/United States
- R380R/1114/DMT_/The Dunhill Medical Trust/United Kingdom
- R01 AG031287/AG/NIA NIH HHS/United States
- U01 AG049505/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

