Exploring QSAR models for activity-cliff prediction
- PMID: 37069675
- PMCID: PMC10107580
- DOI: 10.1186/s13321-023-00708-w
Exploring QSAR models for activity-cliff prediction
Abstract
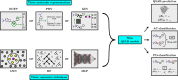
Introduction and methodology: Pairs of similar compounds that only differ by a small structural modification but exhibit a large difference in their binding affinity for a given target are known as activity cliffs (ACs). It has been hypothesised that QSAR models struggle to predict ACs and that ACs thus form a major source of prediction error. However, the AC-prediction power of modern QSAR methods and its quantitative relationship to general QSAR-prediction performance is still underexplored. We systematically construct nine distinct QSAR models by combining three molecular representation methods (extended-connectivity fingerprints, physicochemical-descriptor vectors and graph isomorphism networks) with three regression techniques (random forests, k-nearest neighbours and multilayer perceptrons); we then use each resulting model to classify pairs of similar compounds as ACs or non-ACs and to predict the activities of individual molecules in three case studies: dopamine receptor D2, factor Xa, and SARS-CoV-2 main protease.
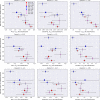
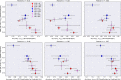
Results and conclusions: Our results provide strong support for the hypothesis that indeed QSAR models frequently fail to predict ACs. We observe low AC-sensitivity amongst the evaluated models when the activities of both compounds are unknown, but a substantial increase in AC-sensitivity when the actual activity of one of the compounds is given. Graph isomorphism features are found to be competitive with or superior to classical molecular representations for AC-classification and can thus be employed as baseline AC-prediction models or simple compound-optimisation tools. For general QSAR-prediction, however, extended-connectivity fingerprints still consistently deliver the best performance amongs the tested input representations. A potential future pathway to improve QSAR-modelling performance might be the development of techniques to increase AC-sensitivity.
Keywords: Activity cliff prediction; Activity cliffs; Binding affinity prediction; Deep learning; Extended-connectivity fingerprints; Graph isomorphism networks; Machine learning; Molecular representation; Physicochemical descriptors; QSAR modelling.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Achdout H, Aimon A, Bar-David E, Barr H, Ben-Shmuel A, Bennett J, Bilenko VA, Bilenko VA, Boby ML, Borden B, Bowman GR, Brun J, et al (2022) Open science discovery of oral non-covalent SARS-CoV-2 main protease inhibitor therapeutics. BioRxiv. https://www.biorxiv.org/content/early/2022/01/30/2020.10.29.339317. Accessed 19 Jan 2023
-
- Akiba T, Sano S, Yanase T, Ohta T, Koyama M (2019) Optuna: a next-generation hyperparameter optimization framework. In: Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, pp 2623–2631
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

