Deciphering molecular mechanisms of SARS-CoV-2 pathogenesis and drug repurposing through GRN motifs: a comprehensive systems biology study
- PMID: 37070032
- PMCID: PMC10090260
- DOI: 10.1007/s13205-023-03518-x
Deciphering molecular mechanisms of SARS-CoV-2 pathogenesis and drug repurposing through GRN motifs: a comprehensive systems biology study
Abstract
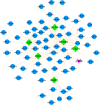
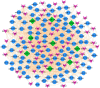
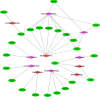
The world has recently been plagued by a new coronavirus infection called SARS-CoV-2. This virus may lead to severe acute respiratory syndrome followed by multiple organ failure. SARS-CoV-2 has approximately 80-90% genetic similarity to SARS-CoV. Given the limited omics data available for host response to the viruses (more limited data for SARS-CoV-2), we attempted to unveil the crucial molecular mechanisms underlying the SARS-CoV-2 pathogenesis by comparing its regulatory network motifs with SARS-CoV. We also attempted to identify the non-shared crucial molecules and their functions to predict the specific mechanisms for each infection and the processes responsible for their different manifestations. Deciphering the crucial shared and non-shared mechanisms at the molecular level and signaling pathways underlying both diseases may help shed light on their pathogenesis and pave the way for other new drug repurposing against COVID-19. We constructed the GRNs for host response to SARS-CoV and SARS-CoV-2 pathogens (in vitro) and identified the significant 3-node regulatory motifs by analyzing them topologically and functionally. We attempted to identify the shared and non-shared regulatory elements and signaling pathways between their host responses. Interestingly, our findings indicated that NFKB1, JUN, STAT1, FOS, KLF4, and EGR1 were the critical shared TFs between motif-related subnetworks in both SARS and COVID-1, which are considered genes with specific functions in the immune response. Enrichment analysis revealed that the NOD-like receptor signaling, TNF signaling, and influenza A pathway were among the first significant pathways shared between SARS and COVID-19 up-regulated DEGs networks, and the term "metabolic pathways" (hsa01100) among the down-regulated DEGs networks. WEE1, PMAIP1, and TSC22D2 were identified as the top three hubs specific to SARS. However, MYPN, SPRY4, and APOL6 were the tops specific to COVID-19 in vitro. The term "Complement and coagulation cascades" pathway was identified as the first top non-shared pathway for COVID-19 and the MAPK signaling pathway for SARS. We used the identified crucial DEGs to construct a drug-gene interaction network to propose some drug candidates. Zinc chloride, Fostamatinib, Copper, Tirofiban, Tretinoin, and Levocarnitine were the six drugs with higher scores in our drug-gene network analysis.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-023-03518-x.
Keywords: COVID-19; DEGs; Host response; Regulatory network motifs; SARS.
© King Abdulaziz City for Science and Technology 2023, Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe authors declare that they have no competing interests.
Figures


Similar articles
-
Deciphering the similarities and disparities of molecular mechanisms behind respiratory epithelium response to HCoV-229E and SARS-CoV-2 and drug repurposing, a systems biology approach.Daru. 2024 Jun;32(1):215-235. doi: 10.1007/s40199-024-00507-0. Epub 2024 Apr 23. Daru. 2024. PMID: 38652363 Free PMC article.
-
Identification of host transcriptome-guided repurposable drugs for SARS-CoV-1 infections and their validation with SARS-CoV-2 infections by using the integrated bioinformatics approaches.PLoS One. 2022 Apr 7;17(4):e0266124. doi: 10.1371/journal.pone.0266124. eCollection 2022. PLoS One. 2022. PMID: 35390032 Free PMC article.
-
Integrative transcriptomics analysis of lung epithelial cells and identification of repurposable drug candidates for COVID-19.Eur J Pharmacol. 2020 Nov 15;887:173594. doi: 10.1016/j.ejphar.2020.173594. Epub 2020 Sep 22. Eur J Pharmacol. 2020. PMID: 32971089 Free PMC article.
-
Coronavirus virulence genes with main focus on SARS-CoV envelope gene.Virus Res. 2014 Dec 19;194:124-37. doi: 10.1016/j.virusres.2014.07.024. Epub 2014 Aug 2. Virus Res. 2014. PMID: 25093995 Free PMC article. Review.
-
Integration of Omics Data and Network Models to Unveil Negative Aspects of SARS-CoV-2, from Pathogenic Mechanisms to Drug Repurposing.Biology (Basel). 2023 Aug 31;12(9):1196. doi: 10.3390/biology12091196. Biology (Basel). 2023. PMID: 37759595 Free PMC article. Review.
Cited by
-
Expression of DDSR1 Long Non-Coding RNA and Genes Involved in the DNA Damage Response in Sperm with DNA Fragmentation.Reprod Sci. 2024 Oct;31(10):3112-3121. doi: 10.1007/s43032-024-01640-6. Epub 2024 Jul 16. Reprod Sci. 2024. PMID: 39014289
-
COVID-19: A novel holistic systems biology approach to predict its molecular mechanisms (in vitro) and repurpose drugs.Daru. 2023 Dec;31(2):155-171. doi: 10.1007/s40199-023-00471-1. Epub 2023 Aug 19. Daru. 2023. PMID: 37597114 Free PMC article.
-
Deciphering the similarities and disparities of molecular mechanisms behind respiratory epithelium response to HCoV-229E and SARS-CoV-2 and drug repurposing, a systems biology approach.Daru. 2024 Jun;32(1):215-235. doi: 10.1007/s40199-024-00507-0. Epub 2024 Apr 23. Daru. 2024. PMID: 38652363 Free PMC article.
-
Deciphering Molecular Mechanisms of Cutaneous Leishmaniasis, Pathogenesis and Drug Repurposing through Systems Biology.Iran Biomed J. 2024 Jul 1;28(4):179-91. doi: 10.61186/ibj.4177. Iran Biomed J. 2024. PMID: 39036455 Free PMC article.
References
-
- Aevermann BD, Pickett BE, Kumar S, Klem EB, Agnihothram S, Askovich PS, Bankhead A, 3rd, Bolles M, Carter V, Chang J, Clauss TR, Dash P, Diercks AH, Eisfeld AJ, Ellis A, Fan S, Ferris MT, Gralinski LE, Green RR, Gritsenko MA, Hatta M, Heegel RA, Jacobs JM, Jeng S, Josset L, Kaiser SM, Kelly S, Law GL, Li C, Li J, Long C, Luna ML, Matzke M, McDermott J, Menachery V, Metz TO, Mitchell H, Monroe ME, Navarro G, Neumann G, Podyminogin RL, Purvine SO, Rosenberger CM, Sanders CJ, Schepmoes AA, Shukla AK, Sims A, Sova P, Tam VC, Tchitchek N, Thomas PG, Tilton SC, Totura A, Wang J, Webb-Robertson BJ, Wen J, Weiss JM, Yang F, Yount B, Zhang Q, McWeeney S, Smith RD, Waters KM, Kawaoka Y, Baric R, Aderem A, Katze MG, Scheuermann RH. A comprehensive collection of systems biology data characterizing the host response to viral infection. Sci Data. 2014;1:140033. - PMC - PubMed
-
- Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, Gris D, Roney KE, Zimmermann AG, Bowzard JB, Ranjan P, Monroe KM, Pickles RJ, Sambhara S, Ting JP. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity. 2011;34:854–865. - PMC - PubMed
-
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

