A simulation-based evaluation of methods for estimating census population size of terrestrial game species from genetically-identified parent-offspring pairs
- PMID: 37070094
- PMCID: PMC10105560
- DOI: 10.7717/peerj.15151
A simulation-based evaluation of methods for estimating census population size of terrestrial game species from genetically-identified parent-offspring pairs
Abstract
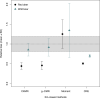
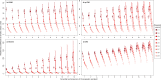
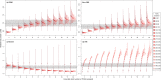
Estimates of wildlife population size are critical for conservation and management, but accurate estimates are difficult to obtain for many species. Several methods have recently been developed that estimate abundance using kinship relationships observed in genetic samples, particularly parent-offspring pairs. While these methods are similar to traditional Capture-Mark-Recapture, they do not need physical recapture, as individuals are considered recaptured if a sample contains one or more close relatives. This makes methods based on genetically-identified parent-offspring pairs particularly interesting for species for which releasing marked animals back into the population is not desirable or not possible (e.g., harvested fish or game species). However, while these methods have successfully been applied in commercially important fish species, in the absence of life-history data, they are making several assumptions unlikely to be met for harvested terrestrial species. They assume that a sample contains only one generation of parents and one generation of juveniles of the year, while more than two generations can coexist in the hunting bags of long-lived species, or that the sampling probability is the same for each individual, an assumption that is violated when fecundity and/or survival depend on sex or other individual traits. In order to assess the usefulness of kin-based methods to estimate population sizes of terrestrial game species, we simulated population pedigrees of two different species with contrasting demographic strategies (wild boar and red deer), applied four different methods and compared the accuracy and precision of their estimates. We also performed a sensitivity analysis, simulating population pedigrees with varying fecundity characteristics and various levels of harvesting to identify optimal conditions of applicability of each method. We showed that all these methods reached the required levels of accuracy and precision to be effective in wildlife management under simulated circumstances (i.e., for species within a given range of fecundity and for a given range of sampling intensity), while being robust to fecundity variation. Despite the potential usefulness of the methods for terrestrial game species, care is needed as several biases linked to hunting practices still need to be investigated (e.g., when hunting bags are biased toward a particular group of individuals).
Keywords: Fecundity; Genetic markers; Kinship; Mark-recapture; Pedigree; Population monitoring; Wildlife management.
©2023 Larroque and Balkenhol.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


Similar articles
-
Assessing the suitability of a one-time sampling event for close-kin mark-recapture: A caribou case study.Ecol Evol. 2024 Sep 3;14(9):e70230. doi: 10.1002/ece3.70230. eCollection 2024 Sep. Ecol Evol. 2024. PMID: 39234160 Free PMC article.
-
Sources of bias in applying close-kin mark-recapture to terrestrial game species with different life histories.Ecology. 2024 Mar;105(3):e4244. doi: 10.1002/ecy.4244. Epub 2024 Jan 25. Ecology. 2024. PMID: 38272487
-
Population size estimates based on the frequency of genetically assigned parent-offspring pairs within a subsample.Ecol Evol. 2020 May 20;10(13):6356-6363. doi: 10.1002/ece3.6365. eCollection 2020 Jul. Ecol Evol. 2020. PMID: 32724517 Free PMC article.
-
Non-invasive genetic censusing and monitoring of primate populations.Am J Primatol. 2018 Mar;80(3):e22743. doi: 10.1002/ajp.22743. Epub 2018 Feb 19. Am J Primatol. 2018. PMID: 29457631 Review.
-
Estimating animal population density using passive acoustics.Biol Rev Camb Philos Soc. 2013 May;88(2):287-309. doi: 10.1111/brv.12001. Epub 2012 Nov 29. Biol Rev Camb Philos Soc. 2013. PMID: 23190144 Free PMC article. Review.
Cited by
-
Advances in wildlife abundance estimation using pedigree reconstruction.Ecol Evol. 2023 Oct 18;13(10):e10650. doi: 10.1002/ece3.10650. eCollection 2023 Oct. Ecol Evol. 2023. PMID: 37869434 Free PMC article.
-
Unraveling the Complexity of the N e/N c Ratio for Conservation of Large and Widespread Pelagic Fish Species: Current Status and Challenges.Evol Appl. 2024 Oct 10;17(10):e70020. doi: 10.1111/eva.70020. eCollection 2024 Oct. Evol Appl. 2024. PMID: 39391864 Free PMC article. Review.
-
Assessing the suitability of a one-time sampling event for close-kin mark-recapture: A caribou case study.Ecol Evol. 2024 Sep 3;14(9):e70230. doi: 10.1002/ece3.70230. eCollection 2024 Sep. Ecol Evol. 2024. PMID: 39234160 Free PMC article.
References
-
- Apollonio M, Andersen R, Putman R. Ungulate management in Europe in the XXI century. Cambridge University Press; Cambridge: 2010.
-
- Bassi E, Gazzola A, Bongi P, Scandura M, Apollonio M. Relative impact of human harvest and wolf predation on two ungulate species in Central Italy. Ecological Research. 2020;35:662–674. doi: 10.1111/1440-1703.12130. - DOI
-
- Blouin MS. DNA-based methods for pedigree reconstruction and kinship analysis in natural populations. Trends in Ecology & Evolution. 2003;18:503–511. doi: 10.1016/S0169-5347(03)00225-8. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

