Saccharomyces cerevisiae DNA polymerase IV overcomes Rad51 inhibition of DNA polymerase δ in Rad52-mediated direct-repeat recombination
- PMID: 37070185
- PMCID: PMC10287921
- DOI: 10.1093/nar/gkad281
Saccharomyces cerevisiae DNA polymerase IV overcomes Rad51 inhibition of DNA polymerase δ in Rad52-mediated direct-repeat recombination
Abstract
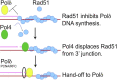
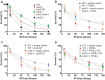
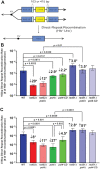
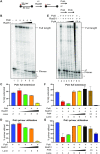
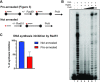
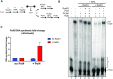
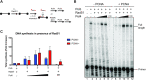
Saccharomyces cerevisiae DNA polymerase IV (Pol4) like its homolog, human DNA polymerase lambda (Polλ), is involved in Non-Homologous End-Joining and Microhomology-Mediated Repair. Using genetic analysis, we identified an additional role of Pol4 also in homology-directed DNA repair, specifically in Rad52-dependent/Rad51-independent direct-repeat recombination. Our results reveal that the requirement for Pol4 in repeat recombination was suppressed by the absence of Rad51, suggesting that Pol4 counteracts the Rad51 inhibition of Rad52-mediated repeat recombination events. Using purified proteins and model substrates, we reconstituted in vitro reactions emulating DNA synthesis during direct-repeat recombination and show that Rad51 directly inhibits Polδ DNA synthesis. Interestingly, although Pol4 was not capable of performing extensive DNA synthesis by itself, it aided Polδ in overcoming the DNA synthesis inhibition by Rad51. In addition, Pol4 dependency and stimulation of Polδ DNA synthesis in the presence of Rad51 occurred in reactions containing Rad52 and RPA where DNA strand-annealing was necessary. Mechanistically, yeast Pol4 displaces Rad51 from ssDNA independent of DNA synthesis. Together our in vitro and in vivo data suggest that Rad51 suppresses Rad52-dependent/Rad51-independent direct-repeat recombination by binding to the primer-template and that Rad51 removal by Pol4 is critical for strand-annealing dependent DNA synthesis.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Interaction of yeast Rad51 and Rad52 relieves Rad52-mediated inhibition of de novo telomere addition.PLoS Genet. 2020 Feb 3;16(2):e1008608. doi: 10.1371/journal.pgen.1008608. eCollection 2020 Feb. PLoS Genet. 2020. PMID: 32012161 Free PMC article.
-
Dynamic regulatory interactions of rad51, rad52, and replication protein-a in recombination intermediates.J Mol Biol. 2009 Jul 3;390(1):45-55. doi: 10.1016/j.jmb.2009.05.009. Epub 2009 May 13. J Mol Biol. 2009. PMID: 19445949
-
Rad51 protein controls Rad52-mediated DNA annealing.J Biol Chem. 2008 May 23;283(21):14883-92. doi: 10.1074/jbc.M801097200. Epub 2008 Mar 12. J Biol Chem. 2008. PMID: 18337252 Free PMC article.
-
Emerging non-canonical roles for the Rad51-Rad52 interaction in response to double-strand breaks in yeast.Curr Genet. 2020 Oct;66(5):917-926. doi: 10.1007/s00294-020-01081-z. Epub 2020 May 12. Curr Genet. 2020. PMID: 32399607 Free PMC article. Review.
-
Non-Recombinogenic Functions of Rad51, BRCA2, and Rad52 in DNA Damage Tolerance.Genes (Basel). 2021 Sep 29;12(10):1550. doi: 10.3390/genes12101550. Genes (Basel). 2021. PMID: 34680945 Free PMC article. Review.
Cited by
-
Cohesin complex oligomerization maintains end-tethering at DNA double-strand breaks.Nat Cell Biol. 2025 Jan;27(1):118-129. doi: 10.1038/s41556-024-01552-2. Epub 2024 Oct 31. Nat Cell Biol. 2025. PMID: 39482358 Free PMC article.
-
Gross Chromosomal Rearrangement at Centromeres.Biomolecules. 2023 Dec 24;14(1):28. doi: 10.3390/biom14010028. Biomolecules. 2023. PMID: 38254628 Free PMC article. Review.
-
CRISPR/Cas9-induced double-strand breaks in the huntingtin locus lead to CAG repeat contraction through DNA end resection and homology-mediated repair.BMC Biol. 2024 Dec 3;22(1):282. doi: 10.1186/s12915-024-02079-6. BMC Biol. 2024. PMID: 39627841 Free PMC article.
-
DNA polymerase ζ is a robust reverse transcriptase.bioRxiv [Preprint]. 2024 Sep 28:2024.09.27.615452. doi: 10.1101/2024.09.27.615452. bioRxiv. 2024. Update in: J Biol Chem. 2024 Dec;300(12):107918. doi: 10.1016/j.jbc.2024.107918. PMID: 39386629 Free PMC article. Updated. Preprint.
References
-
- Kowalczykowski S.C., Hunter N., Heyer W.-D.. DNA Recombination. 2016; Cold Spring Harbor Laboratory Press.
-
- Krogh B.O., Symington L.S.. Recombination proteins in yeast. Annu. Rev. Genet. 2004; 38:233–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

