Interaction of the La-related protein Slf1 with colliding ribosomes maintains translation of oxidative-stress responsive mRNAs
- PMID: 37070186
- PMCID: PMC10287931
- DOI: 10.1093/nar/gkad272
Interaction of the La-related protein Slf1 with colliding ribosomes maintains translation of oxidative-stress responsive mRNAs
Abstract
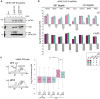
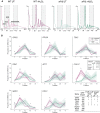
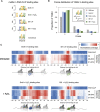
In response to oxidative stress cells reprogram gene expression to enhance levels of antioxidant enzymes and promote survival. In Saccharomyces cerevisiae the polysome-interacting La-related proteins (LARPs) Slf1 and Sro9 aid adaptation of protein synthesis during stress by undetermined means. To gain insight in their mechanisms of action in stress responses, we determined LARP mRNA binding positions in stressed and unstressed cells. Both proteins bind within coding regions of stress-regulated antioxidant enzyme and other highly translated mRNAs in both optimal and stressed conditions. LARP interaction sites are framed and enriched with ribosome footprints suggesting ribosome-LARP-mRNA complexes are identified. Although stress-induced translation of antioxidant enzyme mRNAs is attenuated in slf1Δ, these mRNAs remain on polysomes. Focusing further on Slf1, we find it binds to both monosomes and disomes following RNase treatment. slf1Δ reduces disome enrichment during stress and alters programmed ribosome frameshifting rates. We propose that Slf1 is a ribosome-associated translational modulator that stabilises stalled/collided ribosomes, prevents ribosome frameshifting and so promotes translation of a set of highly-translated mRNAs that together facilitate cell survival and adaptation to stress.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
The yeast La related protein Slf1p is a key activator of translation during the oxidative stress response.PLoS Genet. 2015 Jan 8;11(1):e1004903. doi: 10.1371/journal.pgen.1004903. eCollection 2015 Jan. PLoS Genet. 2015. PMID: 25569619 Free PMC article.
-
Ribosomes in RNA Granules Are Stalled on mRNA Sequences That Are Consensus Sites for FMRP Association.J Neurosci. 2023 Apr 5;43(14):2440-2459. doi: 10.1523/JNEUROSCI.1002-22.2023. Epub 2023 Feb 27. J Neurosci. 2023. PMID: 36849416 Free PMC article.
-
Genome-wide Survey of Ribosome Collision.Cell Rep. 2020 May 5;31(5):107610. doi: 10.1016/j.celrep.2020.107610. Cell Rep. 2020. PMID: 32375038 Free PMC article.
-
A cellular handbook for collided ribosomes: surveillance pathways and collision types.Curr Genet. 2021 Feb;67(1):19-26. doi: 10.1007/s00294-020-01111-w. Epub 2020 Oct 12. Curr Genet. 2021. PMID: 33044589 Free PMC article. Review.
-
Run, Ribosome, Run: From Compromised Translation to Human Health.Antioxid Redox Signal. 2023 Aug;39(4-6):336-350. doi: 10.1089/ars.2022.0157. Epub 2023 May 17. Antioxid Redox Signal. 2023. PMID: 36825529 Review.
Cited by
-
The central role of translation elongation in response to stress.Biochem Soc Trans. 2023 Jun 28;51(3):959-969. doi: 10.1042/BST20220584. Biochem Soc Trans. 2023. PMID: 37318088 Free PMC article.
-
Paralogous translation factors target distinct mRNAs to differentially regulate tolerance to oxidative stress in yeast.Nucleic Acids Res. 2023 Sep 8;51(16):8820-8835. doi: 10.1093/nar/gkad568. Nucleic Acids Res. 2023. PMID: 37449412 Free PMC article.
-
Is there a localized role for translational quality control?RNA. 2023 Nov;29(11):1623-1643. doi: 10.1261/rna.079683.123. Epub 2023 Aug 15. RNA. 2023. PMID: 37582617 Free PMC article.
-
Translation regulation in response to stress.FEBS J. 2024 Dec;291(23):5102-5122. doi: 10.1111/febs.17076. Epub 2024 Feb 3. FEBS J. 2024. PMID: 38308808 Free PMC article. Review.
-
Protein quality control: from molecular mechanisms to therapeutic intervention-EMBO workshop, May 21-26 2023, Srebreno, Croatia.Cell Stress Chaperones. 2023 Nov;28(6):631-640. doi: 10.1007/s12192-023-01383-4. Epub 2023 Sep 20. Cell Stress Chaperones. 2023. PMID: 37731161 Free PMC article. Review.
References
-
- Shenton D., Smirnova J.B., Selley J.N., Carroll K., Hubbard S.J., Pavitt G.D., Ashe M.P., Grant C.M.. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 2006; 281:29011–29021. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

