Catalytic mechanism and pH dependence of a methyltransferase ribozyme (MTR1) from computational enzymology
- PMID: 37070188
- PMCID: PMC10201425
- DOI: 10.1093/nar/gkad260
Catalytic mechanism and pH dependence of a methyltransferase ribozyme (MTR1) from computational enzymology
Abstract
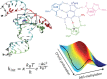
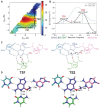
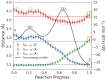
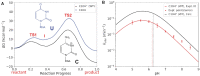
A methyltransferase ribozyme (MTR1) was selected in vitro to catalyze alkyl transfer from exogenous O6-methylguanine (O6mG) to a target adenine N1, and recently, high-resolution crystal structures have become available. We use a combination of classical molecular dynamics, ab initio quantum mechanical/molecular mechanical (QM/MM) and alchemical free energy (AFE) simulations to elucidate the atomic-level solution mechanism of MTR1. Simulations identify an active reactant state involving protonation of C10 that hydrogen bonds with O6mG:N1. The deduced mechanism involves a stepwise mechanism with two transition states corresponding to proton transfer from C10:N3 to O6mG:N1 and rate-controlling methyl transfer (19.4 kcal·mol-1 barrier). AFE simulations predict the pKa for C10 to be 6.3, close to the experimental apparent pKa of 6.2, further implicating it as a critical general acid. The intrinsic rate derived from QM/MM simulations, together with pKa calculations, enables us to predict an activity-pH profile that agrees well with experiment. The insights gained provide further support for a putative RNA world and establish new design principles for RNA-based biochemical tools.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Extensive molecular dynamics simulations showing that canonical G8 and protonated A38H+ forms are most consistent with crystal structures of hairpin ribozyme.J Phys Chem B. 2010 May 20;114(19):6642-52. doi: 10.1021/jp1001258. J Phys Chem B. 2010. PMID: 20420375 Free PMC article.
-
Role of the active site guanine in the glmS ribozyme self-cleavage mechanism: quantum mechanical/molecular mechanical free energy simulations.J Am Chem Soc. 2015 Jan 21;137(2):784-98. doi: 10.1021/ja510387y. Epub 2015 Jan 12. J Am Chem Soc. 2015. PMID: 25526516 Free PMC article.
-
The Role of General Acid Catalysis in the Mechanism of an Alkyl Transferase Ribozyme.ACS Catal. 2024 Oct 2;14(20):15294-15305. doi: 10.1021/acscatal.4c04571. eCollection 2024 Oct 18. ACS Catal. 2024. PMID: 39444533 Free PMC article.
-
Progress in ab initio QM/MM free-energy simulations of electrostatic energies in proteins: accelerated QM/MM studies of pKa, redox reactions and solvation free energies.J Phys Chem B. 2009 Feb 5;113(5):1253-72. doi: 10.1021/jp8071712. J Phys Chem B. 2009. PMID: 19055405 Free PMC article. Review.
-
Catalytic strategies of self-cleaving ribozymes.Acc Chem Res. 2008 Aug;41(8):1027-35. doi: 10.1021/ar800050c. Epub 2008 Jul 25. Acc Chem Res. 2008. PMID: 18652494 Review.
Cited by
-
RNA-Puzzles Round V: blind predictions of 23 RNA structures.Nat Methods. 2025 Feb;22(2):399-411. doi: 10.1038/s41592-024-02543-9. Epub 2024 Dec 2. Nat Methods. 2025. PMID: 39623050 Free PMC article.
-
Electronic and Nuclear Quantum Effects on Proton Transfer Reactions of Guanine-Thymine (G-T) Mispairs Using Combined Quantum Mechanical/Molecular Mechanical and Machine Learning Potentials.Molecules. 2024 Jun 6;29(11):2703. doi: 10.3390/molecules29112703. Molecules. 2024. PMID: 38893576 Free PMC article.
-
Accurately Modeling RNA Stem-Loops in an Implicit Solvent Environment.J Chem Inf Model. 2024 Aug 12;64(15):6092-6104. doi: 10.1021/acs.jcim.4c00756. Epub 2024 Jul 13. J Chem Inf Model. 2024. PMID: 39002142 Free PMC article.
-
Surface-Accelerated String Method for Locating Minimum Free Energy Paths.J Chem Theory Comput. 2024 Mar 12;20(5):2058-2073. doi: 10.1021/acs.jctc.3c01401. Epub 2024 Feb 17. J Chem Theory Comput. 2024. PMID: 38367218 Free PMC article.
-
Structure and catalytic activity of the SAM-utilizing ribozyme SAMURI.Nat Chem Biol. 2025 Jan 8. doi: 10.1038/s41589-024-01808-w. Online ahead of print. Nat Chem Biol. 2025. PMID: 39779902
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

