The autophagy receptor NBR1 directs the clearance of photodamaged chloroplasts
- PMID: 37070813
- PMCID: PMC10156165
- DOI: 10.7554/eLife.86030
The autophagy receptor NBR1 directs the clearance of photodamaged chloroplasts
Abstract
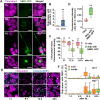
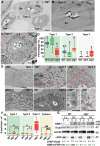
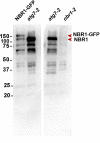
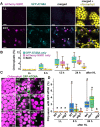
The ubiquitin-binding NBR1 autophagy receptor plays a prominent role in recognizing ubiquitylated protein aggregates for vacuolar degradation by macroautophagy. Here, we show that upon exposing Arabidopsis plants to intense light, NBR1 associates with photodamaged chloroplasts independently of ATG7, a core component of the canonical autophagy machinery. NBR1 coats both the surface and interior of chloroplasts, which is then followed by direct engulfment of the organelles into the central vacuole via a microautophagy-type process. The relocalization of NBR1 into chloroplasts does not require the chloroplast translocon complexes embedded in the envelope but is instead greatly enhanced by removing the self-oligomerization mPB1 domain of NBR1. The delivery of NBR1-decorated chloroplasts into vacuoles depends on the ubiquitin-binding UBA2 domain of NBR1 but is independent of the ubiquitin E3 ligases SP1 and PUB4, known to direct the ubiquitylation of chloroplast surface proteins. Compared to wild-type plants, nbr1 mutants have altered levels of a subset of chloroplast proteins and display abnormal chloroplast density and sizes upon high light exposure. We postulate that, as photodamaged chloroplasts lose envelope integrity, cytosolic ligases reach the chloroplast interior to ubiquitylate thylakoid and stroma proteins which are then recognized by NBR1 for autophagic clearance. This study uncovers a new function of NBR1 in the degradation of damaged chloroplasts by microautophagy.
Keywords: A. thaliana; autophagy receptor; cell biology; chlorophagy; microautophagy; plant biology; ubiquitin.
© 2023, Lee et al.
Conflict of interest statement
HL, JC, AG, KC, JB, SS, AM, RV, KE, MO No competing interests declared
Figures
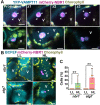
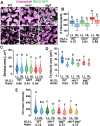
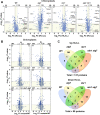


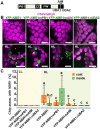
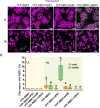
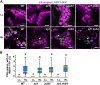
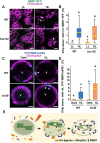
Update of
- doi: 10.1101/2023.01.27.525901
Similar articles
-
NBR1-mediated selective chloroplast autophagy is important to plant stress tolerance.Autophagy. 2024 Jan;20(1):205-206. doi: 10.1080/15548627.2023.2251324. Epub 2023 Aug 27. Autophagy. 2024. PMID: 37635361 Free PMC article.
-
Chlorophagy does not require PLANT U-BOX4-mediated ubiquitination.Plant Signal Behav. 2021 Mar 4;16(3):1861769. doi: 10.1080/15592324.2020.1861769. Epub 2020 Dec 17. Plant Signal Behav. 2021. PMID: 33331806 Free PMC article.
-
The core autophagy machinery is not required for chloroplast singlet oxygen-mediated cell death in the Arabidopsis thaliana plastid ferrochelatase two mutant.BMC Plant Biol. 2021 Jul 19;21(1):342. doi: 10.1186/s12870-021-03119-x. BMC Plant Biol. 2021. PMID: 34281507 Free PMC article.
-
The Role of E3 Ubiquitin Ligases in Chloroplast Function.Int J Mol Sci. 2022 Aug 25;23(17):9613. doi: 10.3390/ijms23179613. Int J Mol Sci. 2022. PMID: 36077009 Free PMC article. Review.
-
The exploitation of host autophagy and ubiquitin machinery by Mycobacterium tuberculosis in shaping immune responses and host defense during infection.Autophagy. 2023 Jan;19(1):3-23. doi: 10.1080/15548627.2021.2021495. Epub 2022 Jan 9. Autophagy. 2023. PMID: 35000542 Free PMC article. Review.
Cited by
-
Genotype-Specific Activation of Autophagy during Heat Wave in Wheat.Cells. 2024 Jul 20;13(14):1226. doi: 10.3390/cells13141226. Cells. 2024. PMID: 39056807 Free PMC article.
-
Autophagosome biogenesis and organelle homeostasis in plant cells.Plant Cell. 2024 Sep 3;36(9):3009-3024. doi: 10.1093/plcell/koae099. Plant Cell. 2024. PMID: 38536783 Free PMC article. Review.
-
Does the polyubiquitination pathway operate inside intact chloroplasts to remove proteins?Plant Cell. 2024 Sep 3;36(9):2984-2989. doi: 10.1093/plcell/koae104. Plant Cell. 2024. PMID: 38683741 Free PMC article. No abstract available.
-
Vacuolar degradation of plant organelles.Plant Cell. 2024 Sep 3;36(9):3036-3056. doi: 10.1093/plcell/koae128. Plant Cell. 2024. PMID: 38657116 Free PMC article. Review.
-
Can autophagy enhance crop resilience to environmental stress?Philos Trans R Soc Lond B Biol Sci. 2025 May 29;380(1927):20240245. doi: 10.1098/rstb.2024.0245. Epub 2025 May 29. Philos Trans R Soc Lond B Biol Sci. 2025. PMID: 40439312 Free PMC article. Review.
References
-
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. P62/Sqstm1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. The Journal of Cell Biology. 2005;171:603–614. doi: 10.1083/jcb.200507002. - DOI - PMC - PubMed
-
- Cha-Molstad H, Sung KS, Hwang J, Kim KA, Yu JE, Yoo YD, Jang JM, Han DH, Molstad M, Kim JG, Lee YJ, Zakrzewska A, Kim SH, Kim ST, Kim SY, Lee HG, Soung NK, Ahn JS, Ciechanover A, Kim BY, Kwon YT. Amino-Terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nature Cell Biology. 2015;17:917–929. doi: 10.1038/ncb3177. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

