The G118R plus R263K Combination of Integrase Mutations Associated with Dolutegravir-Based Treatment Failure Reduces HIV-1 Replicative Capacity and Integration
- PMID: 37071019
- PMCID: PMC10190594
- DOI: 10.1128/aac.01386-22
The G118R plus R263K Combination of Integrase Mutations Associated with Dolutegravir-Based Treatment Failure Reduces HIV-1 Replicative Capacity and Integration
Abstract
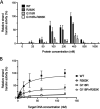
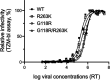
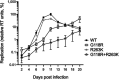
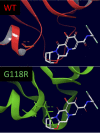
Human immunodeficiency virus (HIV) treatment with antiretroviral regimens containing integrase strand transfer inhibitors such as dolutegravir (DTG) and bictegravir (BIC) offers high levels of protection against the development of drug resistance mutations. Despite this, resistance to DTG and BIC can occur through the development of the R263K integrase substitution. Failure with DTG has also been associated with the emergence of the G118R substitution. G118R and R263K are usually found separately but have been reported together in highly treatment-experienced persons who experienced treatment failure with DTG. We used cell-free strand transfer and DNA binding assays and cell-based infectivity, replicative capacity, and resistance assays to characterize the G118R plus R263K combination of integrase mutations. R263K reduced DTG and BIC susceptibility ~2-fold, in agreement with our previous work. Single-cycle infectivity assays showed that G118R and G118R plus R263K conferred ~10-fold resistance to DTG. G118R alone conferred low levels of resistance to BIC (3.9-fold). However, the G118R plus R263K combination conferred high levels of resistance to BIC (33.7-fold), likely precluding the use of BIC after DTG failure with the G118R plus R263K combination. DNA binding, viral infectivity, and replicative capacity of the double mutant were further impaired, compared to single mutants. We propose that impaired fitness helps to explain the scarcity of the G118R plus R263K combination of integrase substitutions in clinical settings and that immunodeficiency likely contributes to its development.
Keywords: G118R; HIV drug resistance; R263K; bictegravir; dolutegravir; integrase; integrase inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Saag MS, Gandhi RT, Hoy JF, Landovitz RJ, Thompson MA, Sax PE, Smith DM, Benson CA, Buchbinder SP, Del Rio C, Eron JJ, Fätkenheuer G, Günthard HF, Molina J-M, Jacobsen DM, Volberding PA. 2020. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2020 recommendations of the International Antiviral Society-USA Panel. JAMA 324:1651–1669. doi: 10.1001/jama.2020.17025. - DOI - PMC - PubMed
-
- Segal-Maurer S, DeJesus E, Stellbrink H-J, Castagna A, Richmond GJ, Sinclair GI, Siripassorn K, Ruane PJ, Berhe M, Wang H, Margot NA, Dvory-Sobol H, Hyland RH, Brainard DM, Rhee MS, Baeten JM, Molina J-M. CAPELLA Study Investigators. 2022. Capsid inhibition with lenacapavir in multidrug-resistant HIV-1 infection. N Engl J Med 386:1793–1803. doi: 10.1056/NEJMoa2115542. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

