Chiropterans Are a Hotspot for Horizontal Transfer of DNA Transposons in Mammalia
- PMID: 37071810
- PMCID: PMC10162687
- DOI: 10.1093/molbev/msad092
Chiropterans Are a Hotspot for Horizontal Transfer of DNA Transposons in Mammalia
Abstract
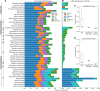
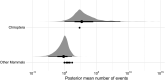
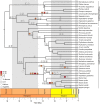
Horizontal transfer of transposable elements (TEs) is an important mechanism contributing to genetic diversity and innovation. Bats (order Chiroptera) have repeatedly been shown to experience horizontal transfer of TEs at what appears to be a high rate compared with other mammals. We investigated the occurrence of horizontally transferred (HT) DNA transposons involving bats. We found over 200 putative HT elements within bats; 16 transposons were shared across distantly related mammalian clades, and 2 other elements were shared with a fish and two lizard species. Our results indicate that bats are a hotspot for horizontal transfer of DNA transposons. These events broadly coincide with the diversification of several bat clades, supporting the hypothesis that DNA transposon invasions have contributed to genetic diversification of bats.
Keywords: echidna; endogenous retrovirus; fusogenic envelope protein; monotremes; platypus.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

References
-
- Amador LI, Moyers Arévalo RL, Almeida FC, Catalano SA, Giannini NP. 2018. Bat systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. J Mammal Evol. 25:37–70.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

