Viral Evolution Shaped by Host Proteostasis Networks
- PMID: 37071930
- PMCID: PMC10543606
- DOI: 10.1146/annurev-virology-100220-112120
Viral Evolution Shaped by Host Proteostasis Networks
Abstract
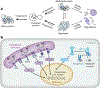
Understanding the factors that shape viral evolution is critical for developing effective antiviral strategies, accurately predicting viral evolution, and preventing pandemics. One fundamental determinant of viral evolution is the interplay between viral protein biophysics and the host machineries that regulate protein folding and quality control. Most adaptive mutations in viruses are biophysically deleterious, resulting in a viral protein product with folding defects. In cells, protein folding is assisted by a dynamic system of chaperones and quality control processes known as the proteostasis network. Host proteostasis networks can determine the fates of viral proteins with biophysical defects, either by assisting with folding or by targeting them for degradation. In this review, we discuss and analyze new discoveries revealing that host proteostasis factors can profoundly shape the sequence space accessible to evolving viral proteins. We also discuss the many opportunities for research progress proffered by the proteostasis perspective on viral evolution and adaptation.
Keywords: chaperone; drug and immune system resistance; protein folding biophysics; quality control; stress response; viral adaptation.
Figures


References
-
- Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet 4:457–69 - PubMed
-
- Worobey M, Holmes EC. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol 80(Part 10):2535–43 - PubMed
-
- Lowen AC. 2017. Constraints, drivers, and implications of influenza A virus reassortment. Annu. Rev. Virol 4:105–21 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

