High-resolution analysis of individual Drosophila melanogaster larvae uncovers individual variability in locomotion and its neurogenetic modulation
- PMID: 37072034
- PMCID: PMC10113034
- DOI: 10.1098/rsob.220308
High-resolution analysis of individual Drosophila melanogaster larvae uncovers individual variability in locomotion and its neurogenetic modulation
Abstract
Neuronally orchestrated muscular movement and locomotion are defining faculties of multicellular animals. Due to its simple brain and genetic accessibility, the larva of the fruit fly Drosophila melanogaster allows one to study these processes at tractable levels of complexity. However, although the faculty of locomotion clearly pertains to the individual, most studies of locomotion in larvae use measurements aggregated across animals, or animals tested one by one, an extravagance for larger-scale analyses. This prevents grasping the inter- and intra-individual variability in locomotion and its neurogenetic determinants. Here, we present the IMBA (individual maggot behaviour analyser) for analysing the behaviour of individual larvae within groups, reliably resolving individual identity across collisions. We use the IMBA to systematically describe the inter- and intra-individual variability in locomotion of wild-type animals, and how the variability is reduced by associative learning. We then report a novel locomotion phenotype of an adhesion GPCR mutant. We further investigated the modulation of locomotion across repeated activations of dopamine neurons in individual animals, and the transient backward locomotion induced by brief optogenetic activation of the brain-descending 'mooncrawler' neurons. In summary, the IMBA is an easy-to-use toolbox allowing an unprecedentedly rich view of the behaviour and its variability of individual larvae, with utility in multiple biomedical research contexts.
Keywords: Latrophilin; dopamine; learning; locomotion; tracking; variability.
Conflict of interest statement
The authors declare no competing interests.
Figures




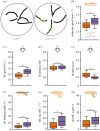






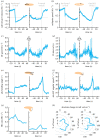
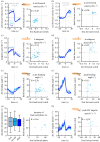

Similar articles
-
The Ol1mpiad: concordance of behavioural faculties of stage 1 and stage 3 Drosophila larvae.J Exp Biol. 2017 Jul 1;220(Pt 13):2452-2475. doi: 10.1242/jeb.156646. J Exp Biol. 2017. PMID: 28679796
-
Potency of transgenic effectors for neurogenetic manipulation in Drosophila larvae.Genetics. 2015 Jan;199(1):25-37. doi: 10.1534/genetics.114.172023. Epub 2014 Oct 29. Genetics. 2015. PMID: 25359929 Free PMC article.
-
Functional Genetic Screen to Identify Interneurons Governing Behaviorally Distinct Aspects of Drosophila Larval Motor Programs.G3 (Bethesda). 2016 Jul 7;6(7):2023-31. doi: 10.1534/g3.116.028472. G3 (Bethesda). 2016. PMID: 27172197 Free PMC article.
-
Transmembrane channel-like (tmc) gene regulates Drosophila larval locomotion.Proc Natl Acad Sci U S A. 2016 Jun 28;113(26):7243-8. doi: 10.1073/pnas.1606537113. Epub 2016 Jun 13. Proc Natl Acad Sci U S A. 2016. PMID: 27298354 Free PMC article.
-
A pair of ascending neurons in the subesophageal zone mediates aversive sensory inputs-evoked backward locomotion in Drosophila larvae.PLoS Genet. 2020 Nov 2;16(11):e1009120. doi: 10.1371/journal.pgen.1009120. eCollection 2020 Nov. PLoS Genet. 2020. PMID: 33137117 Free PMC article.
Cited by
-
Prediction error drives associative learning and conditioned behavior in a spiking model of Drosophila larva.iScience. 2023 Dec 26;27(1):108640. doi: 10.1016/j.isci.2023.108640. eCollection 2024 Jan 19. iScience. 2023. PMID: 38292165 Free PMC article.
-
LarvaTagger: manual and automatic tagging of Drosophila larval behaviour.Bioinformatics. 2024 Jul 1;40(7):btae441. doi: 10.1093/bioinformatics/btae441. Bioinformatics. 2024. PMID: 38970365 Free PMC article.
-
Movement and Dispersion Parameters Characterizing the Group Behavior of Drosophila melanogaster in Micro-Areas of an Observation Arena.Animals (Basel). 2025 May 22;15(11):1515. doi: 10.3390/ani15111515. Animals (Basel). 2025. PMID: 40508981 Free PMC article.
References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

