Low-density lipoprotein receptor-related protein 1 (LRP1) as an auxiliary host factor for RNA viruses
- PMID: 37072184
- PMCID: PMC10114362
- DOI: 10.26508/lsa.202302005
Low-density lipoprotein receptor-related protein 1 (LRP1) as an auxiliary host factor for RNA viruses
Abstract
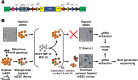
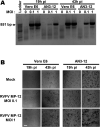
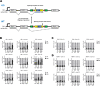
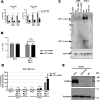
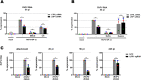
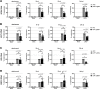
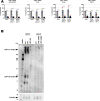
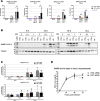
Viruses with an RNA genome are often the cause of zoonotic infections. In order to identify novel pro-viral host cell factors, we screened a haploid insertion-mutagenized mouse embryonic cell library for clones that are resistant to Rift Valley fever virus (RVFV). This screen returned the low-density lipoprotein receptor-related protein 1 (LRP1) as a top hit, a plasma membrane protein involved in a wide variety of cell activities. Inactivation of LRP1 in human cells reduced RVFV RNA levels already at the attachment and entry stages of infection. Moreover, the role of LRP1 in promoting RVFV infection was dependent on physiological levels of cholesterol and on endocytosis. In the human cell line HuH-7, LRP1 also promoted early infection stages of sandfly fever Sicilian virus and La Crosse virus, but had a minor effect on late infection by vesicular stomatitis virus, whereas encephalomyocarditis virus was entirely LRP1-independent. Moreover, siRNA experiments in human Calu-3 cells demonstrated that also SARS-CoV-2 infection benefitted from LRP1. Thus, we identified LRP1 as a host factor that supports infection by a spectrum of RNA viruses.
© 2023 Devignot et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Similar articles
-
Lrp1 is a host entry factor for Rift Valley fever virus.Cell. 2021 Sep 30;184(20):5163-5178.e24. doi: 10.1016/j.cell.2021.09.001. Epub 2021 Sep 23. Cell. 2021. PMID: 34559985 Free PMC article.
-
Lrp1 is essential for lethal Rift Valley fever hepatic disease in mice.Sci Adv. 2023 Jul 14;9(28):eadh2264. doi: 10.1126/sciadv.adh2264. Epub 2023 Jul 14. Sci Adv. 2023. PMID: 37450601 Free PMC article.
-
Host entry factors of Rift Valley Fever Virus infection.Adv Virus Res. 2023;117:121-136. doi: 10.1016/bs.aivir.2023.09.001. Epub 2023 Oct 5. Adv Virus Res. 2023. PMID: 37832991 Free PMC article.
-
Low-density lipoprotein receptor-related protein-1: role in the regulation of vascular integrity.Arterioscler Thromb Vasc Biol. 2014 Mar;34(3):487-98. doi: 10.1161/ATVBAHA.113.301924. Epub 2014 Feb 6. Arterioscler Thromb Vasc Biol. 2014. PMID: 24504736 Free PMC article. Review.
-
Targeting TFIIH to inhibit host cell transcription by Rift Valley Fever Virus.Mol Cell. 2004 Feb 27;13(4):456-8. doi: 10.1016/s1097-2765(04)00092-9. Mol Cell. 2004. PMID: 14992716 Review.
Cited by
-
Crimean-Congo haemorrhagic fever virus uses LDLR to bind and enter host cells.Nat Microbiol. 2024 Jun;9(6):1499-1512. doi: 10.1038/s41564-024-01672-3. Epub 2024 Mar 28. Nat Microbiol. 2024. PMID: 38548922 Free PMC article.
-
The Low-Density Lipoprotein Receptor-Related Protein-1 Is Essential for Dengue Virus Infection.Viruses. 2024 Oct 30;16(11):1692. doi: 10.3390/v16111692. Viruses. 2024. PMID: 39599807 Free PMC article.
-
Lipoprotein receptors: A little grease for enveloped viruses to open the lock?J Biol Chem. 2024 Nov;300(11):107849. doi: 10.1016/j.jbc.2024.107849. Epub 2024 Sep 30. J Biol Chem. 2024. PMID: 39357828 Free PMC article. Review.
-
The LDL receptor-related protein 1 (LRP1) facilitates ACE2-mediated endocytosis of SARS-CoV2 spike protein-containing pseudovirions.J Biol Chem. 2025 Jun;301(6):110227. doi: 10.1016/j.jbc.2025.110227. Epub 2025 May 9. J Biol Chem. 2025. PMID: 40349772 Free PMC article.
-
An Introduction to Rift Valley Fever Virus.Methods Mol Biol. 2024;2824:1-14. doi: 10.1007/978-1-0716-3926-9_1. Methods Mol Biol. 2024. PMID: 39039402
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
