Transcriptomic profiles and 5-year results from the randomized CLL14 study of venetoclax plus obinutuzumab versus chlorambucil plus obinutuzumab in chronic lymphocytic leukemia
- PMID: 37072421
- PMCID: PMC10113251
- DOI: 10.1038/s41467-023-37648-w
Transcriptomic profiles and 5-year results from the randomized CLL14 study of venetoclax plus obinutuzumab versus chlorambucil plus obinutuzumab in chronic lymphocytic leukemia
Erratum in
-
Author Correction: Transcriptomic profiles and 5-year results from the randomized CLL14 study of venetoclax plus obinutuzumab versus chlorambucil plus obinutuzumab in chronic lymphocytic leukemia.Nat Commun. 2023 Oct 23;14(1):6724. doi: 10.1038/s41467-023-42562-2. Nat Commun. 2023. PMID: 37872229 Free PMC article. No abstract available.
Abstract
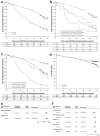
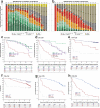
Data on long-term outcomes and biological drivers associated with depth of remission after BCL2 inhibition by venetoclax in the treatment of chronic lymphocytic leukemia (CLL) are limited. In this open-label parallel-group phase-3 study, 432 patients with previously untreated CLL were randomized (1:1) to receive either 1-year venetoclax-obinutuzumab (Ven-Obi, 216 patients) or chlorambucil-Obi (Clb-Obi, 216 patients) therapy (NCT02242942). The primary endpoint was investigator-assessed progression-free survival (PFS); secondary endpoints included minimal residual disease (MRD) and overall survival. RNA sequencing of CD19-enriched blood was conducted for exploratory post-hoc analyses. After a median follow-up of 65.4 months, PFS is significantly superior for Ven-Obi compared to Clb-Obi (Hazard ratio [HR] 0.35 [95% CI 0.26-0.46], p < 0.0001). At 5 years after randomization, the estimated PFS rate is 62.6% after Ven-Obi and 27.0% after Clb-Obi. In both arms, MRD status at the end of therapy is associated with longer PFS. MRD + ( ≥ 10-4) status is associated with increased expression of multi-drug resistance gene ABCB1 (MDR1), whereas MRD6 (< 10-6) is associated with BCL2L11 (BIM) expression. Inflammatory response pathways are enriched in MRD+ patient solely in the Ven-Obi arm. These data indicate sustained long-term efficacy of fixed-duration Ven-Obi in patients with previously untreated CLL. The distinct transcriptomic profile of MRD+ status suggests possible biological vulnerabilities.
© 2023. The Author(s).
Conflict of interest statement
O.A.S.: Advisory Board (Ascentage, AstraZeneca, AbbVie, BeiGene, Eli Lilly, Gilead, Janssen, Roche), speaker honoraria (Adaptive, AstraZeneca, AbbVie, BeiGene, Eli Lilly, Gilead, Janssen, Roche), research funding (BeiGene, AbbVie, Janssen, Roche). H.Y.J.: Employment with Genentech/Hoffmann-la Roche. S.B.: Employment with Genentech/Hoffmann-la Roche. A.K.: Employment with Hoffmann-la Roche. Y.C.: Employment with Genentech/Hoffmann-la Roche. A.M.F.: reports personal fees from Celgene, Janssen, Hoffmann-La Roche. E.T.: Advisory Board (Abbvie, Janssen-Cilag, BeiGene), speaker honoraria (AstraZeneca, Abbvie, BeiGene, Gilead, Janssen, Roche), research funding (Roche, Abbvie, Gilead). C.S.: Speaker honoraria (AstraZeneca, AbbVie). M.R.: reports grants from F.Hoffman-La Roche Ltd; personal fees from F. Hoffmann-La Roche Ltd, AbbVie. K.A.K.: reports grants from F. Hoffmann-La Roche Ltd, Abbvie, during the conduct of the study; personal fees from F. Hoffmann-La Roche Ltd, AbbVie. B.C.: Employment with AbbVie. C.P.P.: Speaker honoraria (AstraZeneca, Pfizer) Research Funding (Gilead Sciences) Advisory Board (Abbvie) J.P.: Employment with Genentech/Hoffmann-la Roche. B.E.: Grants and personal fees from F. Hoffmann-La Roche Ltd, AbbVie, AstraZeneca, BeiGene, and Janssen, personal fees from Celgene, Novartis, ArQule, Gilead, Oxford Biomedica (UK), Adaptive Biotechnologies, Hexal. S.S.: Grants, personal fees and non-financial support from AbbVie, AstraZeneca, Celgene, Gilead, GSK, Hoffmann La-Roche Ltd, Janssen, Novartis, Pharmacyclics, Sunesis, Verastem. Y.J.: Employment with Genentech/Hoffmann-la Roche, stock (Genentech). MH: Honoraria (Roche, Janssen, AbbVie, Gilead Sciences, AstraZeneca), Consulting or Advisory Role (Janssen, AbbVie, Gilead Sciences, Genentech/Roche, AstraZeneca), Speakers’ Bureau: Janssen, AbbVie, Gilead Sciences, Roche/Genentech, AstraZeneca), Research Funding (Roche, AbbVie, Janssen, Gilead Sciences, AstraZeneca), Travel, Accommodations, Expenses (Roche, Janssen). K.F.: Advisory Board (AstraZeneca, AbbVie), speaker honoraria (AbbVie, Roche), research funding (AbbVie, Roche). The remaining authors declare no other competing interests.
Figures


References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA: A Cancer J. Clinicians. 2021;71:7–33. - PubMed
-
- Kater AP, et al. Fixed Duration of Venetoclax-Rituximab in Relapsed/Refractory Chronic Lymphocytic Leukemia Eradicates Minimal Residual Disease and Prolongs Survival: Post-Treatment Follow-Up of the MURANO Phase III Study. J. Clin. Oncol.: Off. J. Am. Soc. Clin. Oncol. 2019;37:269–277. doi: 10.1200/JCO.18.01580. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical

