Plasmepsin X activates the PCRCR complex of Plasmodium falciparum by processing PfRh5 for erythrocyte invasion
- PMID: 37072430
- PMCID: PMC10113190
- DOI: 10.1038/s41467-023-37890-2
Plasmepsin X activates the PCRCR complex of Plasmodium falciparum by processing PfRh5 for erythrocyte invasion
Abstract
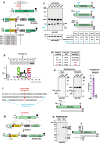
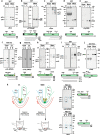
Plasmodium falciparum causes the most severe form of malaria in humans. The protozoan parasite develops within erythrocytes to mature schizonts, that contain more than 16 merozoites, which egress and invade fresh erythrocytes. The aspartic protease plasmepsin X (PMX), processes proteins and proteases essential for merozoite egress from the schizont and invasion of the host erythrocyte, including the leading vaccine candidate PfRh5. PfRh5 is anchored to the merozoite surface through a 5-membered complex (PCRCR), consisting of Plasmodium thrombospondin-related apical merozoite protein, cysteine-rich small secreted protein, Rh5-interacting protein and cysteine-rich protective antigen. Here, we show that PCRCR is processed by PMX in micronemes to remove the N-terminal prodomain of PhRh5 and this activates the function of the complex unmasking a form that can bind basigin on the erythrocyte membrane and mediate merozoite invasion. The ability to activate PCRCR at a specific time in merozoite invasion most likely masks potential deleterious effects of its function until they are required. These results provide an important understanding of the essential role of PMX and the fine regulation of PCRCR function in P. falciparum biology.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- WHO. World Malaria Report 2021 (2021).
-
- Cowman AF, Healer J, Marapana D, Marsh K. Malaria: biology and disease. Cell. 2016;167:610–624. - PubMed
-
- Cowman AF, Tonkin CJ, Tham WH, Duraisingh MT. The molecular basis of erythrocyte invasion by malaria parasites. Cell Host Microbe. 2017;22:232–245. - PubMed
-
- Gilson PR, Crabb BS. Morphology and kinetics of the three distinct phases of red blood cell invasion by Plasmodium falciparum merozoites. Int. J. Parasitol. 2009;39:91–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

