Damage dynamics and the role of chance in the timing of E. coli cell death
- PMID: 37072447
- PMCID: PMC10113371
- DOI: 10.1038/s41467-023-37930-x
Damage dynamics and the role of chance in the timing of E. coli cell death
Abstract
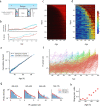
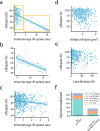
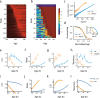
Genetically identical cells in the same stressful condition die at different times. The origin of this stochasticity is unclear; it may arise from different initial conditions that affect the time of demise, or from a stochastic damage accumulation mechanism that erases the initial conditions and instead amplifies noise to generate different lifespans. To address this requires measuring damage dynamics in individual cells over the lifespan, but this has rarely been achieved. Here, we used a microfluidic device to measure membrane damage in 635 carbon-starved Escherichia coli cells at high temporal resolution. We find that initial conditions of damage, size or cell-cycle phase do not explain most of the lifespan variation. Instead, the data points to a stochastic mechanism in which noise is amplified by a rising production of damage that saturates its own removal. Surprisingly, the relative variation in damage drops with age: cells become more similar to each other in terms of relative damage, indicating increasing determinism with age. Thus, chance erases initial conditions and then gives way to increasingly deterministic dynamics that dominate the lifespan distribution.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Finch, C. E. et al. Chance, Development, and Aging. (Oxford University Press, 2000).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

