Definition of the transcriptional units of inherited retinal disease genes by meta-analysis of human retinal transcriptome data
- PMID: 37072692
- PMCID: PMC10111803
- DOI: 10.1186/s12864-023-09300-w
Definition of the transcriptional units of inherited retinal disease genes by meta-analysis of human retinal transcriptome data
Abstract
Background: Inherited retinal diseases (IRD) are genetically heterogeneous disorders that cause the dysfunction or loss of photoreceptor cells and ultimately lead to blindness. To date, next-generation sequencing procedures fail to detect pathogenic sequence variants in coding regions of known IRD disease genes in about 30-40% of patients. One of the possible explanations for this missing heritability is the presence of yet unidentified transcripts of known IRD genes. Here, we aimed to define the transcript composition of IRD genes in the human retina by a meta-analysis of publicly available RNA-seq datasets using an ad-hoc designed pipeline.
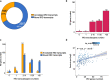
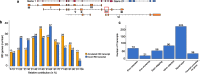
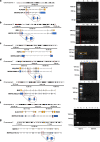
Results: We analysed 218 IRD genes and identified 5,054 transcripts, 3,367 of which were not previously reported. We assessed their putative expression levels and focused our attention on 435 transcripts predicted to account for at least 5% of the expression of the corresponding gene. We looked at the possible impact of the newly identified transcripts at the protein level and experimentally validated a subset of them.
Conclusions: This study provides an unprecedented, detailed overview of the complexity of the human retinal transcriptome that can be instrumental in contributing to the resolution of some cases of missing heritability in IRD patients.
Keywords: Alternative splicing; Human retina; Inherited retinal disease; RNA-seq; Transcriptome.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Where are the missing gene defects in inherited retinal disorders? Intronic and synonymous variants contribute at least to 4% of CACNA1F-mediated inherited retinal disorders.Hum Mutat. 2019 Jun;40(6):765-787. doi: 10.1002/humu.23735. Epub 2019 Mar 28. Hum Mutat. 2019. PMID: 30825406
-
Retinal miRNAs variations in a large cohort of inherited retinal disease.Ophthalmic Genet. 2018 Apr;39(2):175-179. doi: 10.1080/13816810.2017.1329448. Epub 2017 Jul 13. Ophthalmic Genet. 2018. PMID: 28704127
-
The Alter Retina: Alternative Splicing of Retinal Genes in Health and Disease.Int J Mol Sci. 2021 Feb 12;22(4):1855. doi: 10.3390/ijms22041855. Int J Mol Sci. 2021. PMID: 33673358 Free PMC article. Review.
-
Inherited retinal diseases: Linking genes, disease-causing variants, and relevant therapeutic modalities.Prog Retin Eye Res. 2022 Jul;89:101029. doi: 10.1016/j.preteyeres.2021.101029. Epub 2021 Nov 25. Prog Retin Eye Res. 2022. PMID: 34839010 Review.
-
Express: A database of transcriptome profiles encompassing known and novel transcripts across multiple development stages in eye tissues.Exp Eye Res. 2018 Mar;168:57-68. doi: 10.1016/j.exer.2018.01.009. Epub 2018 Jan 11. Exp Eye Res. 2018. PMID: 29337142 Free PMC article.
Cited by
-
Exome sequencing identifies a homozygous splice site variant in RP1 as the underlying cause of autosomal recessive retinitis pigmentosa in a Pakistani family.Ann Med. 2025 Dec;57(1):2470953. doi: 10.1080/07853890.2025.2470953. Epub 2025 Mar 3. Ann Med. 2025. PMID: 40029043 Free PMC article.
-
Deciphering the largest disease-associated transcript isoforms in the human neural retina with advanced long-read sequencing approaches.Genome Res. 2025 Apr 14;35(4):725-739. doi: 10.1101/gr.280060.124. Genome Res. 2025. PMID: 40037841
-
New genetic diagnoses for inherited retinal dystrophies by integrating splicing tools into NGS pipelines.NPJ Genom Med. 2025 Jul 2;10(1):52. doi: 10.1038/s41525-025-00500-9. NPJ Genom Med. 2025. PMID: 40603303 Free PMC article.
-
A proteogenomic atlas of the human neural retina.Front Genet. 2024 Sep 19;15:1451024. doi: 10.3389/fgene.2024.1451024. eCollection 2024. Front Genet. 2024. PMID: 39371417 Free PMC article.
References
-
- Bosze B, Hufnagel RB, Brown NL. Patterning and Cell Type Specification in the Developing CNS and PNS. 2020. Specification of retinal cell types; pp. 481–504.
-
- Daiger SP, Sullivan LS, Bowne SJ. Retinal Information Network (RetNet). https://sph.uth.edu/retnet/. Accessed 20 Jun 2022.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

