Expanding our understanding of marine viral diversity through metagenomic analyses of biofilms
- PMID: 37073293
- PMCID: PMC10077207
- DOI: 10.1007/s42995-020-00078-4
Expanding our understanding of marine viral diversity through metagenomic analyses of biofilms
Abstract
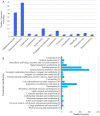
Recent metagenomics surveys have provided insights into the marine virosphere. However, these surveys have focused solely on viruses in seawater, neglecting those associated with biofilms. By analyzing 1.75 terabases of biofilm metagenomic data, 3974 viral sequences were identified from eight locations around the world. Over 90% of these viral sequences were not found in previously reported datasets. Comparisons between biofilm and seawater metagenomes identified viruses that are endemic to the biofilm niche. Analysis of viral sequences integrated within biofilm-derived microbial genomes revealed potential functional genes for trimeric autotransporter adhesin and polysaccharide metabolism, which may contribute to biofilm formation by the bacterial hosts. However, more than 70% of the genes could not be annotated. These findings show marine biofilms to be a reservoir of novel viruses and have enhanced our understanding of natural virus-bacteria ecosystems.
Keywords: Biofilm; Metagenomics; Ocean; Virus.
© Ocean University of China 2021.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures





References
-
- Brum JR, Ignacio-Espinoza JC, Roux S, Doulcier G, Acinas SG, Alberti A, Chaffron S, Cruaud C, De Vargas C, Gasol JM, Gorsky G, Gregory AC, Guidi L, Hingamp P, Iudicone D, Not F, Ogata H, Pesant S, Poulos BT, Schwenck SM, et al. Patterns and ecological drivers of ocean viral communities. Science. 2015;348:1261498. doi: 10.1126/science.1261498. - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources

