The UbiB family member Cqd1 forms a novel membrane contact site in mitochondria
- PMID: 37073556
- PMCID: PMC10214851
- DOI: 10.1242/jcs.260578
The UbiB family member Cqd1 forms a novel membrane contact site in mitochondria
Abstract
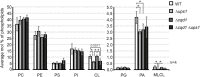
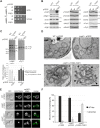
Mitochondria are essential organelles of eukaryotic cells and are characterized by their unique and complex membrane system. They are confined from the cytosol by an envelope consisting of two membranes. Signals, metabolites, proteins and lipids have to be transferred across these membranes via proteinaceous contact sites to keep mitochondria functional. In the present study, we identified a novel mitochondrial contact site in Saccharomyces cerevisiae that is formed by the inner membrane protein Cqd1 and the outer membrane proteins Por1 and Om14. Similar to what is found for the mitochondrial porin Por1, Cqd1 is highly conserved, suggesting that this complex is conserved in form and function from yeast to human. Cqd1 is a member of the UbiB protein kinase-like family (also called aarF domain-containing kinases). It was recently shown that Cqd1, in cooperation with Cqd2, controls the cellular distribution of coenzyme Q by a yet unknown mechanism. Our data suggest that Cqd1 is additionally involved in phospholipid homeostasis. Moreover, overexpression of CQD1 and CQD2 causes tethering of mitochondria to the endoplasmic reticulum, which might explain the ability of Cqd2 to rescue ERMES deletion phenotypes.
Keywords: Contact sites; Mitochondria; Mitochondrial biogenesis; Mitochondrial morphology; Phospholipids; UbiB protein family.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures






Similar articles
-
UbiB proteins regulate cellular CoQ distribution in Saccharomyces cerevisiae.Nat Commun. 2021 Aug 6;12(1):4769. doi: 10.1038/s41467-021-25084-7. Nat Commun. 2021. PMID: 34362905 Free PMC article.
-
An Improved Method to Isolate Mitochondrial Contact Sites.J Vis Exp. 2023 Jun 16;(196). doi: 10.3791/65444. J Vis Exp. 2023. PMID: 37395570
-
Interaction network of the mitochondrial outer membrane protein Mcp3.FEBS Lett. 2018 Oct;592(19):3210-3220. doi: 10.1002/1873-3468.13243. Epub 2018 Sep 29. FEBS Lett. 2018. PMID: 30192984
-
Role of Intra- and Inter-mitochondrial Membrane Contact Sites in Yeast Phospholipid Biogenesis.Adv Exp Med Biol. 2017;997:121-133. doi: 10.1007/978-981-10-4567-7_9. Adv Exp Med Biol. 2017. PMID: 28815526 Review.
-
Phospholipid transport via mitochondria.Traffic. 2014 Sep;15(9):933-45. doi: 10.1111/tra.12188. Epub 2014 Jul 12. Traffic. 2014. PMID: 24954234 Free PMC article. Review.
Cited by
-
New Insights on the Uptake and Trafficking of Coenzyme Q.Antioxidants (Basel). 2023 Jul 6;12(7):1391. doi: 10.3390/antiox12071391. Antioxidants (Basel). 2023. PMID: 37507930 Free PMC article. Review.
-
Nonfunctional coq10 mutants maintain the ERMES complex and reveal true phenotypes associated with the loss of the coenzyme Q chaperone protein Coq10.J Biol Chem. 2024 Nov;300(11):107820. doi: 10.1016/j.jbc.2024.107820. Epub 2024 Sep 27. J Biol Chem. 2024. PMID: 39343004 Free PMC article.
References
-
- Awad, A. M., Nag, A., Pham, N. V. B., Bradley, M. C., Jabassini, N., Nathaniel, J. and Clarke, C. F. (2020). Intragenic suppressor mutations of the COQ8 protein kinase homolog restore coenzyme Q biosynthesis and function in Saccharomyces cerevisiae. PLoS One 15, e0234192. 10.1371/journal.pone.0234192 - DOI - PMC - PubMed
-
- Bohnert, M., Wenz, L. S., Zerbes, R. M., Horvath, S. E., Stroud, D. A., Von Der Malsburg, K., Muller, J. M., Oeljeklaus, S., Perschil, I., Warscheid, B.et al. (2012). Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane. Mol. Biol. Cell 23, 3948-3956. 10.1091/mbc.e12-04-0295 - DOI - PMC - PubMed
-
- Chacinska, A., Rehling, P., Guiard, B., Frazier, A. E., Schulze-Specking, A., Pfanner, N., Voos, W. and Meisinger, C. (2003). Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J. 22, 5370-5381. 10.1093/emboj/cdg532 - DOI - PMC - PubMed
-
- Cherry, J. M., Hong, E. L., Amundsen, C., Balakrishnan, R., Binkley, G., Chan, E. T., Christie, K. R., Costanzo, M. C., Dwight, S. S., Engel, S. R.et al. (2012). Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700-D705. 10.1093/nar/gkr1029 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

