Generation and Functional Analysis of Defective Viral Genomes during SARS-CoV-2 Infection
- PMID: 37074178
- PMCID: PMC10294654
- DOI: 10.1128/mbio.00250-23
Generation and Functional Analysis of Defective Viral Genomes during SARS-CoV-2 Infection
Abstract
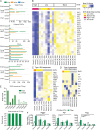
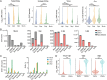
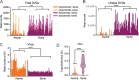
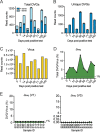
Defective viral genomes (DVGs) have been identified in many RNA viruses as a major factor influencing antiviral immune response and viral pathogenesis. However, the generation and function of DVGs in SARS-CoV-2 infection are less known. In this study, we elucidated DVG generation in SARS-CoV-2 and its relationship with host antiviral immune response. We observed DVGs ubiquitously from transcriptome sequencing (RNA-seq) data sets of in vitro infections and autopsy lung tissues of COVID-19 patients. Four genomic hot spots were identified for DVG recombination, and RNA secondary structures were suggested to mediate DVG formation. Functionally, bulk and single-cell RNA-seq analysis indicated the interferon (IFN) stimulation of SARS-CoV-2 DVGs. We further applied our criteria to the next-generation sequencing (NGS) data set from a published cohort study and observed a significantly higher amount and frequency of DVG in symptomatic patients than those in asymptomatic patients. Finally, we observed exceptionally diverse DVG populations in one immunosuppressive patient up to 140 days after the first positive test of COVID-19, suggesting for the first time an association between DVGs and persistent viral infections in SARS-CoV-2. Together, our findings strongly suggest a critical role of DVGs in modulating host IFN responses and symptom development, calling for further inquiry into the mechanisms of DVG generation and into how DVGs modulate host responses and infection outcome during SARS-CoV-2 infection. IMPORTANCE Defective viral genomes (DVGs) are generated ubiquitously in many RNA viruses, including SARS-CoV-2. Their interference activity to full-length viruses and IFN stimulation provide the potential for them to be used in novel antiviral therapies and vaccine development. SARS-CoV-2 DVGs are generated through the recombination of two discontinuous genomic fragments by viral polymerase complex, and this recombination is also one of the major mechanisms for the emergence of new coronaviruses. Focusing on the generation and function of SARS-CoV-2 DVGs, these studies identify new hot spots for nonhomologous recombination and strongly suggest that the secondary structures within viral genomes mediate the recombination. Furthermore, these studies provide the first evidence for IFN stimulation activity of de novo DVGs during natural SARS-CoV-2 infection. These findings set up the foundation for further mechanism studies of SARS-CoV-2 recombination and provide evidence to harness the immunostimulatory potential of DVGs in the development of a vaccine and antivirals for SARS-CoV-2.
Keywords: RNA secondary structure; SARS-CoV-2; defective viral genomes; human epithelial cells; recombination; secondary structure; type I/III IFN responses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Update of
-
Generation and functional analysis of defective viral genomes during SARS-CoV-2 infection.bioRxiv [Preprint]. 2022 Sep 23:2022.09.22.509123. doi: 10.1101/2022.09.22.509123. bioRxiv. 2022. Update in: mBio. 2023 Jun 27;14(3):e0025023. doi: 10.1128/mbio.00250-23. PMID: 36172120 Free PMC article. Updated. Preprint.
Comment in
-
Bioinformatic analysis of defective viral genomes in SARS-CoV-2 and its impact on population infection characteristics.Front Immunol. 2024 Jan 29;15:1341906. doi: 10.3389/fimmu.2024.1341906. eCollection 2024. Front Immunol. 2024. PMID: 38348041 Free PMC article.
References
-
- Dadras O, Afsahi AM, Pashaei Z, Mojdeganlou H, Karimi A, Habibi P, Barzegary A, Fakhfouri A, Mirzapour P, Janfaza N, Dehghani S, Afroughi F, Dashti M, Khodaei S, Mehraeen E, Voltarelli F, Sabatier JM, SeyedAlinaghi S. 2022. The relationship between COVID-19 viral load and disease severity: a systematic review. Immun Inflamm Dis 10:e580. doi:10.1002/iid3.580. - DOI - PMC - PubMed
-
- Janssen NAF, Grondman I, de Nooijer AH, Boahen CK, Koeken V, Matzaraki V, Kumar V, He X, Kox M, Koenen H, Smeets RL, Joosten I, Brüggemann RJM, Kouijzer IJE, van der Hoeven HG, Schouten JA, Frenzel T, Reijers MHE, Hoefsloot W, Dofferhoff ASM, van Apeldoorn MJ, Blaauw MJT, Veerman K, Maas C, Schoneveld AH, Hoefer IE, Derde LPG, van Deuren M, van der Meer JWM, van Crevel R, Giamarellos-Bourboulis EJ, Joosten LAB, van den Heuvel MM, Hoogerwerf J, de Mast Q, Pickkers P, Netea MG, van de Veerdonk FL. 2021. Dysregulated innate and adaptive immune responses discriminate disease severity in COVID-19. J Infect Dis 223:1322–1333. doi:10.1093/infdis/jiab065. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

