Vibrio cholerae Invasion Dynamics of the Chironomid Host Are Strongly Influenced by Aquatic Cell Density and Can Vary by Strain
- PMID: 37074192
- PMCID: PMC10269514
- DOI: 10.1128/spectrum.02652-22
Vibrio cholerae Invasion Dynamics of the Chironomid Host Are Strongly Influenced by Aquatic Cell Density and Can Vary by Strain
Abstract
Cholera has been a human scourge since the early 1800s and remains a global public health challenge, caused by the toxigenic strains of the bacterium Vibrio cholerae. In its aquatic reservoirs, V. cholerae has been shown to live in association with various arthropod hosts, including the chironomids, a diverse insect family commonly found in wet and semiwet habitats. The association between V. cholerae and chironomids may shield the bacterium from environmental stressors and amplify its dissemination. However, the interaction dynamics between V. cholerae and chironomids remain largely unknown. In this study, we developed freshwater microcosms with chironomid larvae to test the effects of cell density and strain on V. cholerae-chironomid interactions. Our results show that chironomid larvae can be exposed to V. cholerae up to a high inoculation dose (109 cells/mL) without observable detrimental effects. Meanwhile, interstrain variability in host invasion, including prevalence, bacterial load, and effects on host survival, was highly cell density-dependent. Microbiome analysis of the chironomid samples by 16S rRNA gene amplicon sequencing revealed a general effect of V. cholerae exposure on microbiome species evenness. Taken together, our results provide novel insights into V. cholerae invasion dynamics of the chironomid larvae with respect to various doses and strains. The findings suggest that aquatic cell density is a crucial driver of V. cholerae invasion success in chironomid larvae and pave the way for future work examining the effects of a broader dose range and environmental variables (e.g., temperature) on V. cholerae-chironomid interactions. IMPORTANCE Vibrio cholerae is the causative agent of cholera, a significant diarrheal disease affecting millions of people worldwide. Increasing evidence suggests that the environmental facets of the V. cholerae life cycle involve symbiotic associations with aquatic arthropods, which may facilitate its environmental persistence and dissemination. However, the dynamics of interactions between V. cholerae and aquatic arthropods remain unexplored. This study capitalized on using freshwater microcosms with chironomid larvae to investigate the effects of bacterial cell density and strain on V. cholerae-chironomid interactions. Our results suggest that aquatic cell density is the primary determinant of V. cholerae invasion success in chironomid larvae, while interstrain variability in invasion outcomes can be observed under specific cell density conditions. We also determined that V. cholerae exposure generally reduces species evenness of the chironomid-associated microbiome. Collectively, these findings provide novel insights into V. cholerae-arthropod interactions using a newly developed experimental host system.
Keywords: Vibrio cholerae; chironomids; host-microbe interactions; microbiome; microcosm.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

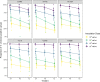
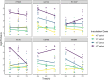
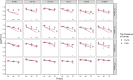
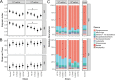
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

