Epigenomic regulation of macrophage polarization: Where do the nuclear receptors belong?
- PMID: 37074820
- PMCID: PMC10524119
- DOI: 10.1111/imr.13209
Epigenomic regulation of macrophage polarization: Where do the nuclear receptors belong?
Abstract
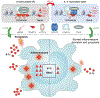
Our laboratory has a long-standing research interest in understanding how lipid-activated transcription factors, nuclear hormone receptors, contribute to dendritic cell and macrophage gene expression regulation, subtype specification, and responses to a changing extra and intracellular milieu. This journey in the last more than two decades took us from identifying target genes for various RXR heterodimers to systematically mapping nuclear receptor-mediated pathways in dendritic cells to identifying hierarchies of transcription factors in alternative polarization in macrophages to broaden the role of nuclear receptors beyond strictly ligand-regulated gene expression. We detail here the milestones of the road traveled and draw conclusions regarding the unexpectedly broad role of nuclear hormone receptors as epigenomic components of dendritic cell and macrophage gene regulation as we are getting ready for the next challenges.
Keywords: EGR2; IL-4; PPARγ; STAT6; epigenome; macrophage; nuclear receptors; polarization; transcriptional regulation.
© 2023 The Authors. Immunological Reviews published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

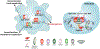





References
-
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of Monocytes, Macrophages, and Dendritic Cells. https://www.science.org - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

